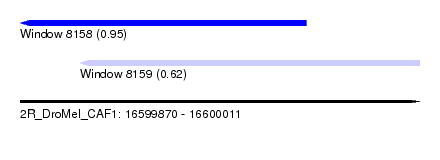
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,599,870 – 16,600,011 |
| Length | 141 |
| Max. P | 0.950543 |

| Location | 16,599,870 – 16,599,971 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -24.82 |
| Energy contribution | -25.44 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
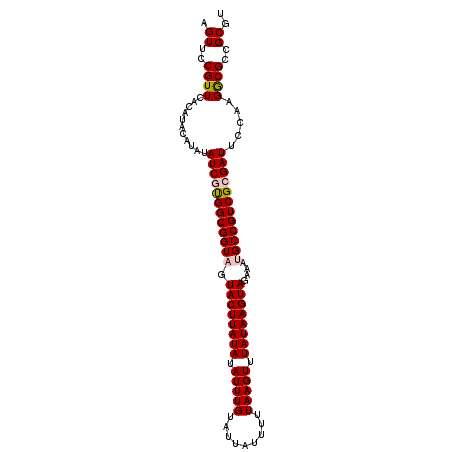
>2R_DroMel_CAF1 16599870 101 - 20766785 AGUUCCGUUCACAUACAUAUAUCGCGGCGGUUGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGUCGCGAUUCCAAGGCGCCGCGU .((.((((......))....((((((((((((.((((((((.(((((.........))))).)))))))).)...))))))))))).....)).))..... ( -29.80) >DroSec_CAF1 7092 101 - 1 AGUUCCGUUCACAUACAUAUAUCUUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGUCGCGAUUCCAAGGCGCCGCGU .((.((((......))....(((.((((((((.((((((((.(((((.........))))).))))))))....)))))))).))).....)).))..... ( -23.10) >DroSim_CAF1 7121 101 - 1 AGUUCCGUUCACAUACAUAUAUCUUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGUCGCGAUUCCAAGGCGCCGCGC .((.((((......))....(((.((((((((.((((((((.(((((.........))))).))))))))....)))))))).))).....)).))..... ( -23.10) >DroYak_CAF1 7103 97 - 1 AGUUCCGUUCGCA----UAUAUCGUGGCGGUUGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGUCGCGAUUCCAAGACGCCGCGU .((..((((....----...((((((((((((.((((((((.(((((.........))))).)))))))).)...))))))))))).....))))..)).. ( -29.50) >consensus AGUUCCGUUCACAUACAUAUAUCGUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGUCGCGAUUCCAAGGCGCCGCGU .((..((((...........((((((((((((.((((((((.(((((.........))))).))))))))....)))))))))))).....))))..)).. (-24.82 = -25.44 + 0.62)

| Location | 16,599,891 – 16,600,011 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -22.70 |
| Energy contribution | -23.51 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16599891 120 - 20766785 UAGGAACUCAAAACAUUGGGCACUUUAUGGUCAUUAUUCAAGUUCCGUUCACAUACAUAUAUCGCGGCGGUUGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGU ..((((((........(((.((.....)).))).......)))))).................((((((.((.((((((((.(((((.........))))).)))))))).)).)))))) ( -29.56) >DroSec_CAF1 7113 120 - 1 CAGGAACUCAAAACAUUGGGCACUUUGUGGUCAUUAUUCAAGUUCCGUUCACAUACAUAUAUCUUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGU ..((((((........(((.(((...))).))).......))))))....................((((((.((((((((.(((((.........))))).))))))))....)))))) ( -27.16) >DroSim_CAF1 7142 120 - 1 CAGGAACUCAAAACAUUGGGCACUUUGUGGUCAUUAUUCAAGUUCCGUUCACAUACAUAUAUCUUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGU ..((((((........(((.(((...))).))).......))))))....................((((((.((((((((.(((((.........))))).))))))))....)))))) ( -27.16) >DroYak_CAF1 7124 116 - 1 CAAGAACUCAAAACAUUGGGCACUUUAUAGUCAUUAUUCAAGUUCCGUUCGCA----UAUAUCGUGGCGGUUGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGU ...((((.(((....)))((.((((.((((...))))..)))).))))))...----........((((.((.((((((((.(((((.........))))).)))))))).)).)))).. ( -22.80) >consensus CAGGAACUCAAAACAUUGGGCACUUUAUGGUCAUUAUUCAAGUUCCGUUCACAUACAUAUAUCGUGGCGGUAGUACUUAUAUAUUUGUAUUAUUUUUAAGUUUAUAAGUAGAAAUGCCGU ..((((((........(((.((.....)).))).......))))))..................(((((.((.((((((((.(((((.........))))).)))))))).)).))))). (-22.70 = -23.51 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:36 2006