| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,595,209 – 16,595,306 |
| Length | 97 |
| Max. P | 0.956912 |

| Location | 16,595,209 – 16,595,306 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 70.82 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16595209 97 + 20766785 AACGA-GCUUCCAGGAAACUGCUCAAAUAACCCUAACUAACCCUUCUAAAAGUGUACAUAUGAAAUGAAA-UUUGG--GGCAGCAGUUUCC--UACGCCUUUG .....-((....(((((((((((.......((((((...((.(((....))).)).(((.....)))...-.))))--)).))))))))))--)..))..... ( -21.90) >DroSec_CAF1 2507 95 + 1 UACGA-GCUUCAAGGAAACUGCUCAAAUG--CCUAACUAACCCUUCUAAAGAUGUACAUAAAAUAUGAAA-UUUGG--GGCAGCAGUUUCC--UACGCCUUUG .....-((....(((((((((((.....(--(((..........((....))....((((...))))...-....)--)))))))))))))--)..))..... ( -21.20) >DroSim_CAF1 2509 95 + 1 UACGA-GCUUCAAGGAAACUGCUCCAAUG--CCUAACUAACCCUUCUAAAGAUGUACAUAUAAUAUGAAA-UUUGG--GGCAGCAGUUUCC--UACGCCUUUG .....-((....(((((((((((.....(--(((..........((....))....((((...))))...-....)--)))))))))))))--)..))..... ( -21.20) >DroEre_CAF1 2527 92 + 1 UACGACGCUUACAGGAAACUGCUCAAAUG--CCCUACUAAACUUCCUGAAAAUGC----AUGAUUUCAAA-UUCCG--GCCAGCAGUUUCC--UACGCUUUUG ......((....(((((((((((.....(--((..........((.((......)----).)).......-....)--)).))))))))))--)..))..... ( -20.20) >DroYak_CAF1 2499 92 + 1 UACGACGCUUCAAGGAAACUGCUCAAAUA--CCACACUACCUCUUCUAAAACUGC----AUGAUAUCAAA-UUCGG--UCCAGCAGUUUCC--UACGCUUUUG ......((....(((((((((((.....(--((...........((.........----..)).......-...))--)..))))))))))--)..))..... ( -17.10) >DroPer_CAF1 11711 88 + 1 AAUGGCUUUCAAGGGUAGCUGCUAAU----------CGAAAUUCACUAAAAA-CC----AAGACAACAAAAUUCGGACUCUAGCAGUAUCCCCUGUGCCUACA ...(((...((.(((..(((((((.(----------((((............-..----............)))))....)))))))..))).)).))).... ( -20.87) >consensus UACGA_GCUUCAAGGAAACUGCUCAAAUG__CCUAACUAACCCUUCUAAAAAUGC____AUGAUAUCAAA_UUCGG__GCCAGCAGUUUCC__UACGCCUUUG ......((.....((((((((((..........................................................)))))))))).....))..... (-11.44 = -11.67 + 0.22)

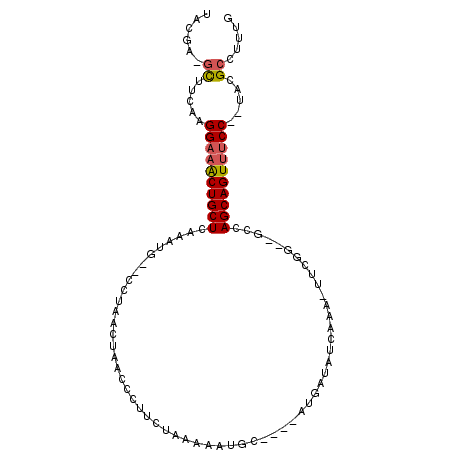
| Location | 16,595,209 – 16,595,306 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 70.82 |
| Mean single sequence MFE | -25.83 |
| Consensus MFE | -11.42 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.717185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16595209 97 - 20766785 CAAAGGCGUA--GGAAACUGCUGCC--CCAAA-UUUCAUUUCAUAUGUACACUUUUAGAAGGGUUAGUUAGGGUUAUUUGAGCAGUUUCCUGGAAGC-UCGUU ....(((.((--(((((((((((((--(.((.-.....((((...............))))......)).))))......))))))))))))...))-).... ( -25.66) >DroSec_CAF1 2507 95 - 1 CAAAGGCGUA--GGAAACUGCUGCC--CCAAA-UUUCAUAUUUUAUGUACAUCUUUAGAAGGGUUAGUUAGG--CAUUUGAGCAGUUUCCUUGAAGC-UCGUA ....(((..(--(((((((((((((--.....-..((...(((((..........))))).)).......))--).....)))))))))))....))-).... ( -23.34) >DroSim_CAF1 2509 95 - 1 CAAAGGCGUA--GGAAACUGCUGCC--CCAAA-UUUCAUAUUAUAUGUACAUCUUUAGAAGGGUUAGUUAGG--CAUUGGAGCAGUUUCCUUGAAGC-UCGUA ....(((..(--((((((((((.((--((...-...((((...))))....((....)).)))...(.....--)...).)))))))))))....))-).... ( -23.60) >DroEre_CAF1 2527 92 - 1 CAAAAGCGUA--GGAAACUGCUGGC--CGGAA-UUUGAAAUCAU----GCAUUUUCAGGAAGUUUAGUAGGG--CAUUUGAGCAGUUUCCUGUAAGCGUCGUA .....((.((--((((((((((.((--(.(((-(((((((....----....)))))...))))).....))--).....))))))))))))...))...... ( -26.80) >DroYak_CAF1 2499 92 - 1 CAAAAGCGUA--GGAAACUGCUGGA--CCGAA-UUUGAUAUCAU----GCAGUUUUAGAAGAGGUAGUGUGG--UAUUUGAGCAGUUUCCUUGAAGCGUCGUA .....((..(--((((((((((.((--.....-....(((((((----((..((((....))))..))))))--))))).)))))))))))....))...... ( -28.50) >DroPer_CAF1 11711 88 - 1 UGUAGGCACAGGGGAUACUGCUAGAGUCCGAAUUUUGUUGUCUU----GG-UUUUUAGUGAAUUUCG----------AUUAGCAGCUACCCUUGAAAGCCAUU ....(((.((((((.(((((((((....((((.((..(((....----..-....)))..)).))))----------.))))))).))))))))...)))... ( -27.10) >consensus CAAAGGCGUA__GGAAACUGCUGCC__CCAAA_UUUCAUAUCAU____ACAUUUUUAGAAGGGUUAGUUAGG__CAUUUGAGCAGUUUCCUUGAAGC_UCGUA .....((.....((((((((((..........................................................)))))))))).....))...... (-11.42 = -12.25 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:34 2006