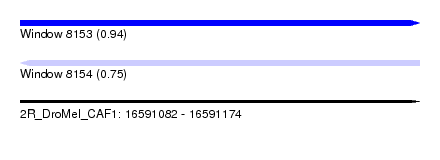
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,591,082 – 16,591,174 |
| Length | 92 |
| Max. P | 0.939262 |

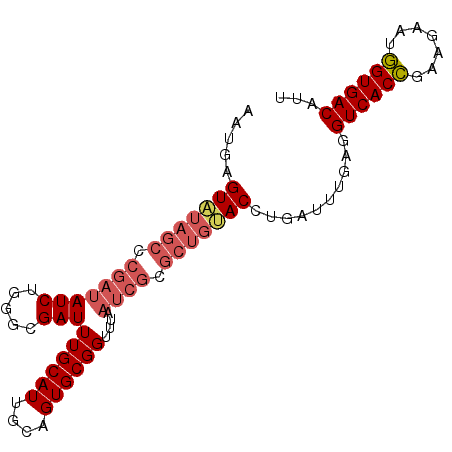
| Location | 16,591,082 – 16,591,174 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -17.30 |
| Energy contribution | -18.18 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16591082 92 + 20766785 AAUGUCACCAUUCUUCGGUGACCUCAAAUCGGGUACAGCGCGAUUAAACCGCACUGCAAUGCAAAUCGCUCAGAUAUCGGGCUAUACUCAUU ...((((((.......))))))........(((((.(((.((((......(((......)))..(((.....))))))).))).)))))... ( -27.10) >DroSec_CAF1 12668 84 + 1 AAUGUCACCAUUCUUCGGUGACCUCAAAUCAGGUACAACGCGAUUAAACCGCACUGCAAUGCAAAUCGGCCAGAU--------AUACUCAUU ...((((((.......)))))).....(((.(((.....(((.......)))..(((...))).....))).)))--------......... ( -16.90) >DroSim_CAF1 12713 92 + 1 AAUGUCACCAUUCUUCGGUGACCUCAAAUCAGGUACAGCCCGAUUAAACCGCACUGCAAUGCAAAUCGCCCAGAUAUCGGGCUAUACUCAUU ...((((((.......)))))).........((((.((((((((......(((......)))..(((.....))))))))))).)))).... ( -28.40) >DroEre_CAF1 12662 92 + 1 AAUGUCACUAUUCUUCAGUGACCUCAAAUCAGGUUCAGCACGAUUAAACCGCACUGCAAUGCAAAUCGCAGAGAUAUCGGGCUCUACUCAUU ...((((((.......)))))).........(((..(((.((((.......(.((((.((....)).)))).)..)))).)))..))).... ( -18.70) >DroYak_CAF1 13056 92 + 1 AAUGUCACUUUUCUUCAGUGACCUCAAAUCAGGUUCAGCGCGAUUAAUCCGCACUGCAAUGCAAAUCGCAGAGAUAUCGGGCUAAACUCAUU ...((((((.......)))))).........((((.(((.((((...((.((..(((...)))....)).))...)))).))).)))).... ( -22.40) >consensus AAUGUCACCAUUCUUCGGUGACCUCAAAUCAGGUACAGCGCGAUUAAACCGCACUGCAAUGCAAAUCGCACAGAUAUCGGGCUAUACUCAUU ...((((((.......)))))).........((((.(((.((((......(((......)))..(((.....))))))).))).)))).... (-17.30 = -18.18 + 0.88)

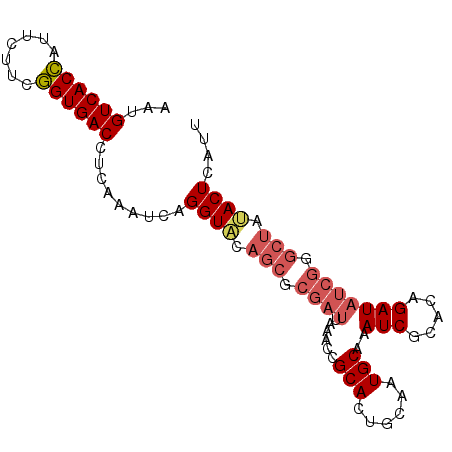
| Location | 16,591,082 – 16,591,174 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.66 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16591082 92 - 20766785 AAUGAGUAUAGCCCGAUAUCUGAGCGAUUUGCAUUGCAGUGCGGUUUAAUCGCGCUGUACCCGAUUUGAGGUCACCGAAGAAUGGUGACAUU ..((.(((((((.((((...(((((....(((((....)))))))))))))).))))))).)).......(((((((.....)))))))... ( -28.40) >DroSec_CAF1 12668 84 - 1 AAUGAGUAU--------AUCUGGCCGAUUUGCAUUGCAGUGCGGUUUAAUCGCGUUGUACCUGAUUUGAGGUCACCGAAGAAUGGUGACAUU .....(((.--------(((.....))).)))..(((((((((((...)))))))))))...........(((((((.....)))))))... ( -21.70) >DroSim_CAF1 12713 92 - 1 AAUGAGUAUAGCCCGAUAUCUGGGCGAUUUGCAUUGCAGUGCGGUUUAAUCGGGCUGUACCUGAUUUGAGGUCACCGAAGAAUGGUGACAUU .....((((((((((((..(((.(((((....)))))....)))....))))))))))))..........(((((((.....)))))))... ( -34.40) >DroEre_CAF1 12662 92 - 1 AAUGAGUAGAGCCCGAUAUCUCUGCGAUUUGCAUUGCAGUGCGGUUUAAUCGUGCUGAACCUGAUUUGAGGUCACUGAAGAAUAGUGACAUU .....((((((........))))))...........(((..((((...))))..))).............(((((((.....)))))))... ( -24.30) >DroYak_CAF1 13056 92 - 1 AAUGAGUUUAGCCCGAUAUCUCUGCGAUUUGCAUUGCAGUGCGGAUUAAUCGCGCUGAACCUGAUUUGAGGUCACUGAAGAAAAGUGACAUU .....(((((((.((((...((((((...(((...))).))))))...)))).)))))))..........((((((.......))))))... ( -29.60) >consensus AAUGAGUAUAGCCCGAUAUCUGGGCGAUUUGCAUUGCAGUGCGGUUUAAUCGCGCUGUACCUGAUUUGAGGUCACCGAAGAAUGGUGACAUU .....(((((((.(((((((.....)))((((((....))))))....)))).)))))))..........((((((.......))))))... (-20.58 = -21.66 + 1.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:31 2006