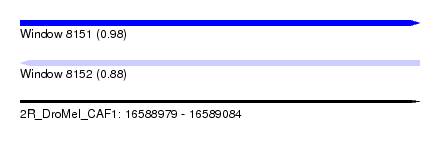
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,588,979 – 16,589,084 |
| Length | 105 |
| Max. P | 0.979402 |

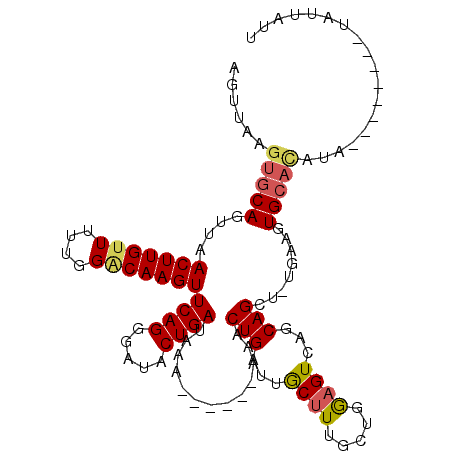
| Location | 16,588,979 – 16,589,084 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -10.68 |
| Energy contribution | -10.92 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979402 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16588979 105 + 20766785 U-UUCUA--------UAUGUGCACUUCA-AGCUGCUGACUUCAACGAGGCAAUCAGUUUUUUUUUUUUAUCAGUAUCCCUGAACUUGUCCAAAAACAAGUUAAGUGCACUUAACU .-.....--------...((((((((.(-(((((.((.((((...))))))..))))))..........((((.....))))((((((......)))))).))))))))...... ( -24.00) >DroSec_CAF1 10625 103 + 1 AAUAAUA--------UAUGUGCACUUCAAAGCUGCUGACUCCAGCAAAGCAAUCAGUUUU----UUUUAUCAGUAUCCCUGAACUUGUCCAAAAACAAGUUAACUGCACUUAACU .......--------...((((........(((((((....))))..)))....((((..----.....((((.....))))((((((......)))))).))))))))...... ( -21.30) >DroSim_CAF1 10679 101 + 1 AAUAAUA--------UAUGUGCACUUCAAAGCUGCUGACUCCAGCAAAGCAAUCAGUUU------UUUAUCAGUAUCCCUGAACUUGUCCAAAAACAAGUUAACUGCACUCAACU .......--------...((((........(((((((....))))..)))....((((.------....((((.....))))((((((......)))))).))))))))...... ( -21.30) >DroEre_CAF1 10681 108 + 1 AAAAAUACAUACAAGUAUAAGCACUUGA-AUCUGGUUACUCCUGGAAAGUAAUCAGUUA------UUUAUCAGUACCACUGAACUUGUCCAAGAUCAAGUUUACUGUGGCAAAGU .....(((...(((((......))))).-..(((((((((.......)))))))))...------.......)))((((((((((((........))))))))..))))...... ( -25.10) >DroYak_CAF1 11002 100 + 1 CAAAAUA--------UAUAUGCACUUGA-ACCUGCUGACUCCUGCAAAGUAAUCAGUAA------UUUAUCAGUAUCACUGAACUUGCCCGAAAACAAGUUUACUGCACUAAAGU .......--------....((((.((((-...((((((...((....))...)))))).------....))))......((((((((........)))))))).))))....... ( -15.40) >consensus AAUAAUA________UAUGUGCACUUCA_AGCUGCUGACUCCAGCAAAGCAAUCAGUUU______UUUAUCAGUAUCCCUGAACUUGUCCAAAAACAAGUUAACUGCACUAAACU ..................(((((.........((((...........))))..................((((.....))))(((((........)))))....)))))...... (-10.68 = -10.92 + 0.24)

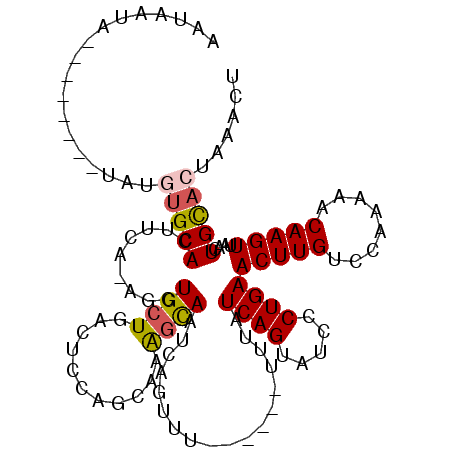
| Location | 16,588,979 – 16,589,084 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.48 |
| Mean single sequence MFE | -25.22 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16588979 105 - 20766785 AGUUAAGUGCACUUAACUUGUUUUUGGACAAGUUCAGGGAUACUGAUAAAAAAAAAAAACUGAUUGCCUCGUUGAAGUCAGCAGCU-UGAAGUGCACAUA--------UAGAA-A ......((((((((.(((((((....)))))))((((.....)))).............((((((..(.....).)))))).....-..))))))))...--------.....-. ( -26.40) >DroSec_CAF1 10625 103 - 1 AGUUAAGUGCAGUUAACUUGUUUUUGGACAAGUUCAGGGAUACUGAUAAAA----AAAACUGAUUGCUUUGCUGGAGUCAGCAGCUUUGAAGUGCACAUA--------UAUUAUU ......(((((.((.(((((((....)))))))((((.....)))).....----....((((((.(......).))))))........)).)))))...--------....... ( -25.10) >DroSim_CAF1 10679 101 - 1 AGUUGAGUGCAGUUAACUUGUUUUUGGACAAGUUCAGGGAUACUGAUAAA------AAACUGAUUGCUUUGCUGGAGUCAGCAGCUUUGAAGUGCACAUA--------UAUUAUU ......(((((.((.(((((((....)))))))((((.....))))....------...((((((.(......).))))))........)).)))))...--------....... ( -25.10) >DroEre_CAF1 10681 108 - 1 ACUUUGCCACAGUAAACUUGAUCUUGGACAAGUUCAGUGGUACUGAUAAA------UAACUGAUUACUUUCCAGGAGUAACCAGAU-UCAAGUGCUUAUACUUGUAUGUAUUUUU .(..((((((....((((((........))))))..))))))..).....------...(((.((((((.....)))))).)))..-.((((((....))))))........... ( -22.80) >DroYak_CAF1 11002 100 - 1 ACUUUAGUGCAGUAAACUUGUUUUCGGGCAAGUUCAGUGAUACUGAUAAA------UUACUGAUUACUUUGCAGGAGUCAGCAGGU-UCAAGUGCAUAUA--------UAUUUUG ......(((((.(.(((((((((((..((((((((((((((........)------)))))))..)).))))..)))..)))))))-)..).)))))...--------....... ( -26.70) >consensus AGUUAAGUGCAGUUAACUUGUUUUUGGACAAGUUCAGGGAUACUGAUAAA______AAACUGAUUGCUUUGCUGGAGUCAGCAGCU_UGAAGUGCACAUA________UAUUAUU ......(((((....(((((((....)))))))((((.....)))).............(((...((((.....))))...)))........))))).................. (-16.76 = -17.12 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:29 2006