
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,588,564 – 16,588,678 |
| Length | 114 |
| Max. P | 0.598538 |

| Location | 16,588,564 – 16,588,678 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -18.27 |
| Energy contribution | -23.02 |
| Covariance contribution | 4.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16588564 114 + 20766785 UC-CCUCAUAUCUGCUGAUAUUUUCCCUGAGCUUUUUUUAUCGCGUUGAAAACGUGAAUUCUCGUUAUUUUUACCACAGAUCAGCGACUUGCGGUGUCAGUGAUCUGUGGUCCCU ..-.((((((((....)))).......)))).........((((((.....))))))...............(((((((((((..(((.......)))..))))))))))).... ( -29.91) >DroSim_CAF1 10292 110 + 1 UC-CCUCAUAUCAGCUGAUCUUUUCCCUGAGCUU----UAUCGCGCUGAAAACGUGAAUUCUCGUUCAUUUUACCACAGAUCAGCGACUUGCGGUGUCAGUGAUCUGUGGUCCCC ..-.......((((((((((((......)))...----.)))).)))))....((((((....))))))...(((((((((((..(((.......)))..))))))))))).... ( -31.90) >DroEre_CAF1 10295 104 + 1 UC-CCUGAUAUCUGCUGAUAUUUUCCCUGAGCUU----UAUAGCGUUGAAAACGUGGUUUCGCGUAUUUUUUAC------UCACUGACUUGCGGUGUCAGUGAUCUGUGGUCACC ..-...((((((....))))))..(((.(((((.----...)))((.(((.(((((....))))).)))...))------((((((((.......)))))))))).).))..... ( -24.50) >DroYak_CAF1 10605 105 + 1 UCCCCUGAUAUCUGCUGAUAUUUUCCCUGAGCUU----UAUCUCGUUGAAAACGUGGAUUAGCGUUAUUUUUAC------UCACUGACUUGCAGUGUCAGUGAUCUGUGGUCUCC ......((((((....))))))..(((.(((((.----.(((((((.....))).)))).)))...........------((((((((.......)))))))))).).))..... ( -24.00) >consensus UC_CCUCAUAUCUGCUGAUAUUUUCCCUGAGCUU____UAUCGCGUUGAAAACGUGAAUUCGCGUUAUUUUUAC______UCACCGACUUGCGGUGUCAGUGAUCUGUGGUCCCC .......(((((....)))))...................((((((.....))))))...............((((((((((((((((.......)))))))))))))))).... (-18.27 = -23.02 + 4.75)

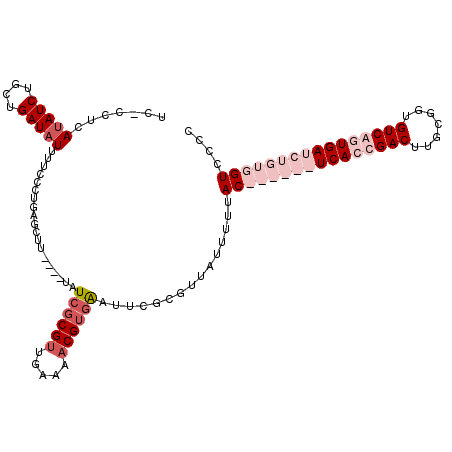

| Location | 16,588,564 – 16,588,678 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.75 |
| Mean single sequence MFE | -26.69 |
| Consensus MFE | -18.99 |
| Energy contribution | -22.43 |
| Covariance contribution | 3.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16588564 114 - 20766785 AGGGACCACAGAUCACUGACACCGCAAGUCGCUGAUCUGUGGUAAAAAUAACGAGAAUUCACGUUUUCAACGCGAUAAAAAAAGCUCAGGGAAAAUAUCAGCAGAUAUGAGG-GA .((((((((((((((.((((.......)))).))))))))))).........((((((....))))))................))).......(((((....)))))....-.. ( -30.10) >DroSim_CAF1 10292 110 - 1 GGGGACCACAGAUCACUGACACCGCAAGUCGCUGAUCUGUGGUAAAAUGAACGAGAAUUCACGUUUUCAGCGCGAUA----AAGCUCAGGGAAAAGAUCAGCUGAUAUGAGG-GA ....(((((((((((.((((.......)))).))))))))))).....(((((.(....).)))))(((((((....----..)).....((.....)).))))).......-.. ( -33.30) >DroEre_CAF1 10295 104 - 1 GGUGACCACAGAUCACUGACACCGCAAGUCAGUGA------GUAAAAAAUACGCGAAACCACGUUUUCAACGCUAUA----AAGCUCAGGGAAAAUAUCAGCAGAUAUCAGG-GA .(((........((((((((.......))))))))------..........)))....((...(((((...(((...----.)))....)))))(((((....)))))..))-.. ( -20.97) >DroYak_CAF1 10605 105 - 1 GGAGACCACAGAUCACUGACACUGCAAGUCAGUGA------GUAAAAAUAACGCUAAUCCACGUUUUCAACGAGAUA----AAGCUCAGGGAAAAUAUCAGCAGAUAUCAGGGGA .....((.....((((((((.......))))))))------...........(((.((....((((((...(((...----...)))...)))))))).)))........))... ( -22.40) >consensus GGGGACCACAGAUCACUGACACCGCAAGUCACUGA______GUAAAAAUAACGAGAAUCCACGUUUUCAACGCGAUA____AAGCUCAGGGAAAAUAUCAGCAGAUAUCAGG_GA .(((((((((((((((((((.......))))))))))))))))..................(((.....)))............))).......(((((....)))))....... (-18.99 = -22.43 + 3.44)

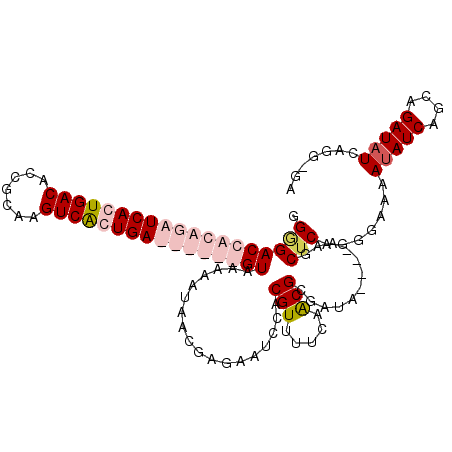
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:27 2006