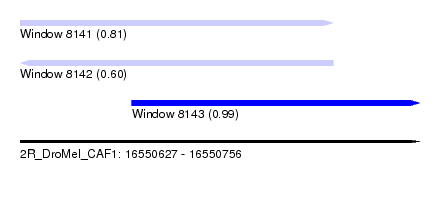
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,550,627 – 16,550,756 |
| Length | 129 |
| Max. P | 0.986315 |

| Location | 16,550,627 – 16,550,728 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -19.56 |
| Energy contribution | -19.45 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
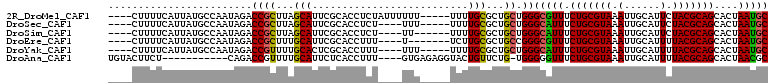
>2R_DroMel_CAF1 16550627 101 + 20766785 GCAUUAGUGCUGCGUAGAAUGCAAUUUACGCAGAAACGCCCAGCAGCGCAAAA-----AAAAAAUAGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAG---- .(((((...(((((((((......)))))))))...(((..((((.((((...-----............))))..))))..)))(((....))).))))).....---- ( -28.06) >DroSec_CAF1 17399 97 + 1 GCAUUAGUGCUGCGUAGAAUGCAAUUUACGCAGAAAUGCCCAGCAGCGCAAAA-----AAA----AGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAG---- .(((((((((((((((((......)))))(((....)))...)))))))....-----...----...........((((((........))))))))))).....---- ( -28.60) >DroSim_CAF1 20121 96 + 1 GCAUUAGUGCUGCGUAGAAUGCAAUUUACGCAGAAAUGCCCAGCAGCGCAAAA------AA----AGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAG---- .(((((((((((((((((......)))))(((....)))...)))))))....------..----...........((((((........))))))))))).....---- ( -28.60) >DroEre_CAF1 17490 95 + 1 GCAUUAGUGCUGCGUAAAAUGCAAUUUACGCAGAAACGCCCGGCAGCGCAAGA-------A----AAAGGUGCGAAUGCAAAGCGGUCUAUUGGCAUAAUGAAAAG---- (((((....(((((((((......)))))))))...(((((.....(....).-------.----...)).))))))))......(((....)))...........---- ( -28.30) >DroYak_CAF1 18398 97 + 1 GCAUUAGUGCUGCGUAAAAUGCAAUUUACGCAGAAAUGCCCAGCAGCGCAAAA-----AAA----AAAGGUGCGAGUGCAAAACGGUCUAUUGGCAUAAUGAAAAG---- .((((.((((((((((((......)))))(((....)))...)))))))....-----...----....((((.((((..........)))).)))))))).....---- ( -26.80) >DroAna_CAF1 17207 94 + 1 GCGUUAGUGCUGCGUAAAAUGCAAUUUACGCAGAAACCCCCA-CAGAACAGUACCUCUCAC----AAAGGUGAGAAUGCAAAACGGUCUG-----------AGAAGUACA ......((((((((((((......)))))))...........-.....(((.(((((((((----....)))))).........))))))-----------...))))). ( -24.90) >consensus GCAUUAGUGCUGCGUAAAAUGCAAUUUACGCAGAAACGCCCAGCAGCGCAAAA_____AAA____AAAGGUGCGAAUGCAAAGCGGUCUAUUGGCAUAAUGAAAAG____ (((((....(((((((((......)))))))))...(((((...........................)).))))))))......(((....)))............... (-19.56 = -19.45 + -0.11)

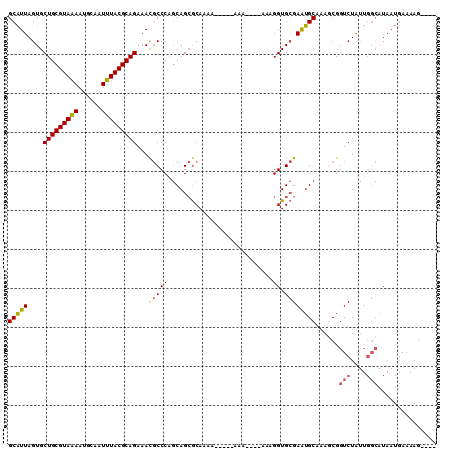
| Location | 16,550,627 – 16,550,728 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.61 |
| Mean single sequence MFE | -26.68 |
| Consensus MFE | -14.89 |
| Energy contribution | -14.47 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.600983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16550627 101 - 20766785 ----CUUUUCAUUAUGCCAAUAGACCGCUUAGCAUUCGCACCUCUAUUUUUU-----UUUUGCGCUGCUGGGCGUUUCUGCGUAAAUUGCAUUCUACGCAGCACUAAUGC ----.....((((((((....(((.(((((((((..((((............-----...)))).)))))))))..)))(((((..........))))).))).))))). ( -27.16) >DroSec_CAF1 17399 97 - 1 ----CUUUUCAUUAUGCCAAUAGACCGCUUAGCAUUCGCACCUCU----UUU-----UUUUGCGCUGCUGGGCAUUUCUGCGUAAAUUGCAUUCUACGCAGCACUAAUGC ----.....((((((((.........((((((((..((((.....----...-----...)))).)))))))).....((((((..........))))))))).))))). ( -26.00) >DroSim_CAF1 20121 96 - 1 ----CUUUUCAUUAUGCCAAUAGACCGCUUAGCAUUCGCACCUCU----UU------UUUUGCGCUGCUGGGCAUUUCUGCGUAAAUUGCAUUCUACGCAGCACUAAUGC ----.....((((((((.........((((((((..((((.....----..------...)))).)))))))).....((((((..........))))))))).))))). ( -26.10) >DroEre_CAF1 17490 95 - 1 ----CUUUUCAUUAUGCCAAUAGACCGCUUUGCAUUCGCACCUUU----U-------UCUUGCGCUGCCGGGCGUUUCUGCGUAAAUUGCAUUUUACGCAGCACUAAUGC ----.....((((((((....(((.((((..(((..((((.....----.-------...)))).)))..))))..)))(((((((......))))))).))).))))). ( -27.30) >DroYak_CAF1 18398 97 - 1 ----CUUUUCAUUAUGCCAAUAGACCGUUUUGCACUCGCACCUUU----UUU-----UUUUGCGCUGCUGGGCAUUUCUGCGUAAAUUGCAUUUUACGCAGCACUAAUGC ----.....((((((((.........(((..(((..((((.....----...-----...)))).)))..))).....((((((((......))))))))))).))))). ( -23.20) >DroAna_CAF1 17207 94 - 1 UGUACUUCU-----------CAGACCGUUUUGCAUUCUCACCUUU----GUGAGAGGUACUGUUCUG-UGGGGGUUUCUGCGUAAAUUGCAUUUUACGCAGCACUAACGC ....((((.-----------((((.((...(((.(((((((....----)))))))))).)).))))-.))))(((.(((((((((......)))))))))....))).. ( -30.30) >consensus ____CUUUUCAUUAUGCCAAUAGACCGCUUAGCAUUCGCACCUCU____UUU_____UUUUGCGCUGCUGGGCAUUUCUGCGUAAAUUGCAUUCUACGCAGCACUAAUGC ........................((((...(((..........................)))...)).))(((((.(((((((.(......).)))))))....))))) (-14.89 = -14.47 + -0.42)
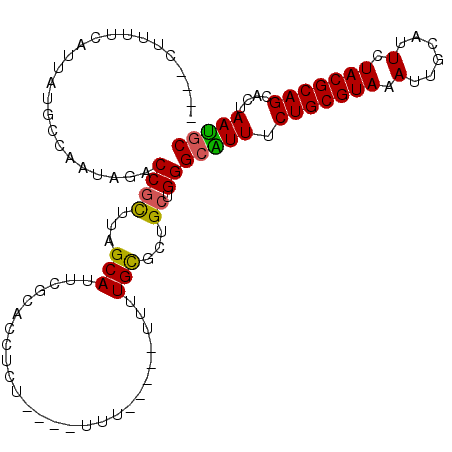
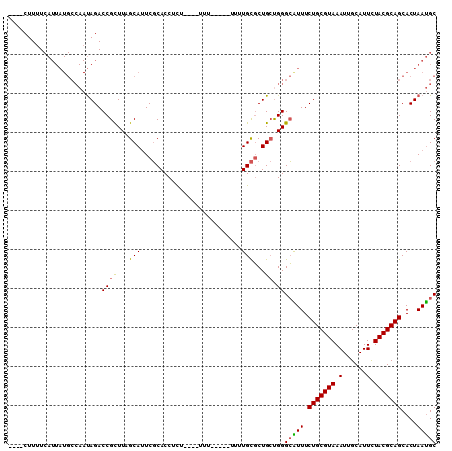
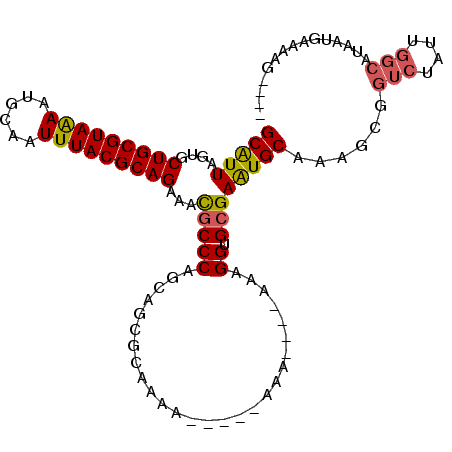
| Location | 16,550,663 – 16,550,756 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -21.48 |
| Energy contribution | -20.96 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986315 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16550663 93 + 20766785 CGCCCAGCAGCGCAAAAAAAAAAUAGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAGCAGUAACAAUGCAGCAGCAACACCUUGA (((......))).............((((((....((((.....(((....)))..........(((.......)))))))....)))))).. ( -19.60) >DroSec_CAF1 17435 89 + 1 UGCCCAGCAGCGCAAAAAAA----AGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAGCGGUAACAAUGCAGCAGCAGCACCUUGA (((.(....).)))......----.(((((((..(((((((........)))))))........((.(((....))).))....))))))).. ( -23.10) >DroSim_CAF1 20157 88 + 1 UGCCCAGCAGCGCAAAA-AA----AGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAGCGGUAACAAUGCAGCAGCAGCACCUUGA (((.(....).)))...-..----.(((((((..(((((((........)))))))........((.(((....))).))....))))))).. ( -23.10) >DroEre_CAF1 17526 87 + 1 CGCCCGGCAGCGCAAGA--A----AAAGGUGCGAAUGCAAAGCGGUCUAUUGGCAUAAUGAAAAGCAGUAAUAAUGCAGCAGCAGCACCUUGA (((......))).....--.----.(((((((...(((......(((....)))..........(((.......))).)))...))))))).. ( -23.30) >DroYak_CAF1 18434 89 + 1 UGCCCAGCAGCGCAAAAAAA----AAAGGUGCGAGUGCAAAACGGUCUAUUGGCAUAAUGAAAAGCAGUAAUAAUGCAGCAGCAGCACCUUGA (((.(....).)))......----.(((((((...(((......(((....)))..........(((.......))).)))...))))))).. ( -21.60) >consensus UGCCCAGCAGCGCAAAAAAA____AGAGGUGCGAAUGCUAAGCGGUCUAUUGGCAUAAUGAAAAGCAGUAACAAUGCAGCAGCAGCACCUUGA (((......))).............(((((((...(((......(((....)))..........(((.......))).)))...))))))).. (-21.48 = -20.96 + -0.52)

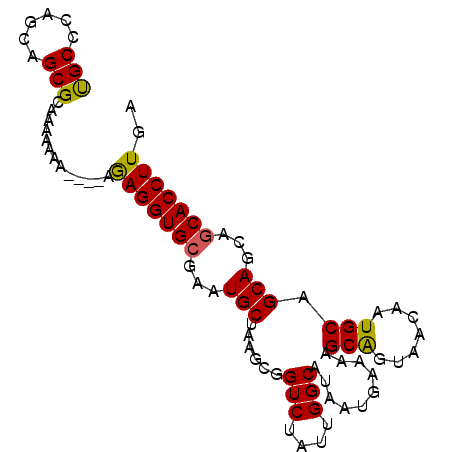
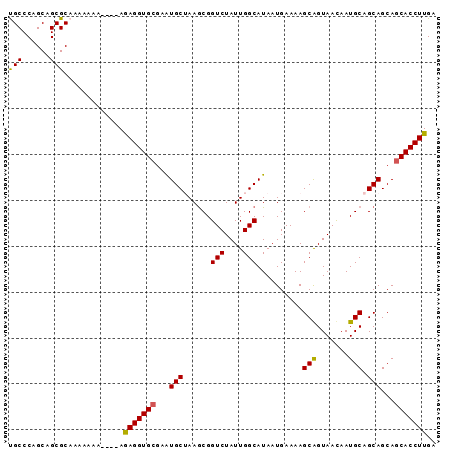
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:21 2006