
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,536,329 – 16,536,425 |
| Length | 96 |
| Max. P | 0.876491 |

| Location | 16,536,329 – 16,536,425 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -18.69 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
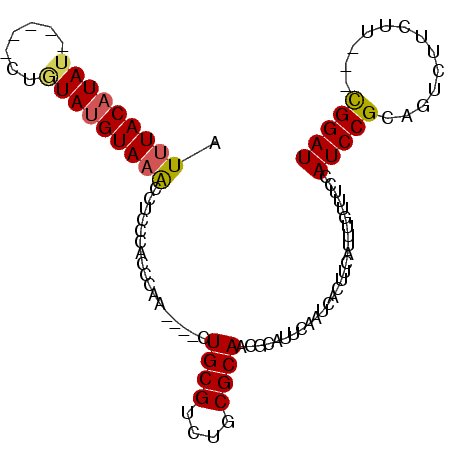
>2R_DroMel_CAF1 16536329 96 + 20766785 AUCCGAGGAAGAAGACGGCGGAUGGAAACAAAUGAAGUGAUUGAAUGCGUUGCGCAGACGCAG----UUGGGUGGGAGGUUUACAUAUAG-----AUAUGUAAAU ((((((........((..((..((....))..))..)).......((((((.....)))))).----)))))).....(((((((((...-----.))))))))) ( -23.10) >DroSec_CAF1 3371 93 + 1 AUCCG---AAGAAGACGGCGGAUGGAAACAAAUGAAGUGAUUGAAUGCGUUGCGCAGACGCAG----UUGGGUGGGAGGUUUACAUACAG-----AUAUGUAAAU (((((---(.....((..((..((....))..))..)).......((((((.....)))))).----)))))).....(((((((((...-----.))))))))) ( -22.90) >DroEre_CAF1 3373 96 + 1 AUCCG---AAGAAGACUGCGGAUGGAAACAAAUGAAGUGAUUGAAUGCGUUGCGCAGACGCAGUCAGUCGGGUGGGAGGCUUACGUACU------AUAUGUAAAU (((((---(....(((((((..((....))...............((((...))))..)))))))..)))))).......(((((((..------.))))))).. ( -28.40) >DroYak_CAF1 3403 98 + 1 AUCCA---AAGAAGACUGCGGAUGGAAACAAAUGAAGUGAUUGAAUGCGUUGCGCAGACGCAG----UUGGGUGGGAGGUAUACAUACAUAUUUCUUAUGUAAAU .((((---.....(((((((..((....))...............((((...))))..)))))----))...)))).........((((((.....))))))... ( -25.10) >consensus AUCCG___AAGAAGACGGCGGAUGGAAACAAAUGAAGUGAUUGAAUGCGUUGCGCAGACGCAG____UUGGGUGGGAGGUUUACAUACAG_____AUAUGUAAAU (((((.........(((.((..((....))..)).))).......((((((.....))))))......))))).....(((((((((.........))))))))) (-18.69 = -19.12 + 0.44)


| Location | 16,536,329 – 16,536,425 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -15.07 |
| Consensus MFE | -10.94 |
| Energy contribution | -11.20 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16536329 96 - 20766785 AUUUACAUAU-----CUAUAUGUAAACCUCCCACCCAA----CUGCGUCUGCGCAACGCAUUCAAUCACUUCAUUUGUUUCCAUCCGCCGUCUUCUUCCUCGGAU .((((((((.-----...))))))))............----.((((....))))...........................(((((..(.......)..))))) ( -12.90) >DroSec_CAF1 3371 93 - 1 AUUUACAUAU-----CUGUAUGUAAACCUCCCACCCAA----CUGCGUCUGCGCAACGCAUUCAAUCACUUCAUUUGUUUCCAUCCGCCGUCUUCUU---CGGAU .((((((((.-----...))))))))............----.((((....))))...........................(((((..(....)..---))))) ( -13.70) >DroEre_CAF1 3373 96 - 1 AUUUACAUAU------AGUACGUAAGCCUCCCACCCGACUGACUGCGUCUGCGCAACGCAUUCAAUCACUUCAUUUGUUUCCAUCCGCAGUCUUCUU---CGGAU .(((((....------.....)))))........((((..(((((((..((.(.((((.................)))).)))..)))))))....)---))).. ( -19.03) >DroYak_CAF1 3403 98 - 1 AUUUACAUAAGAAAUAUGUAUGUAUACCUCCCACCCAA----CUGCGUCUGCGCAACGCAUUCAAUCACUUCAUUUGUUUCCAUCCGCAGUCUUCUU---UGGAU ...((((((.....))))))........(((......(----(((((..((.(.((((.................)))).)))..))))))......---.))). ( -14.65) >consensus AUUUACAUAU_____CUGUAUGUAAACCUCCCACCCAA____CUGCGUCUGCGCAACGCAUUCAAUCACUUCAUUUGUUUCCAUCCGCAGUCUUCUU___CGGAU .(((((((((.......))))))))).................((((....))))...........................(((((.............))))) (-10.94 = -11.20 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:14 2006