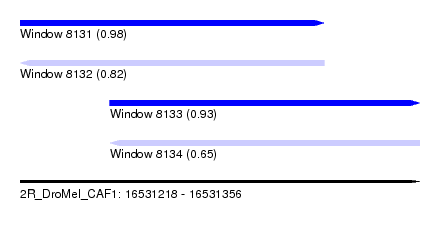
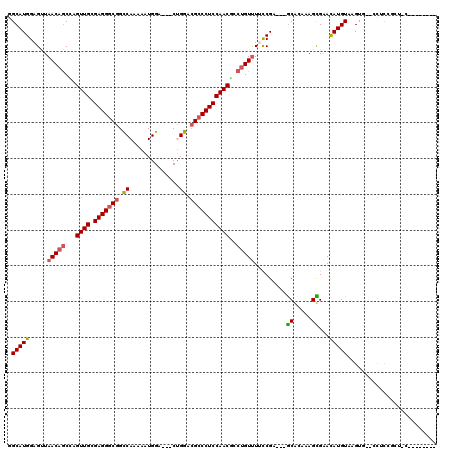
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,531,218 – 16,531,356 |
| Length | 138 |
| Max. P | 0.977369 |

| Location | 16,531,218 – 16,531,323 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -28.43 |
| Energy contribution | -28.74 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
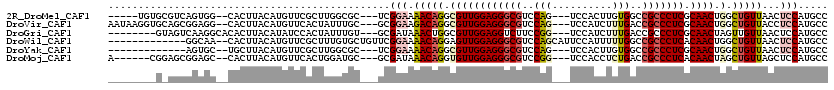
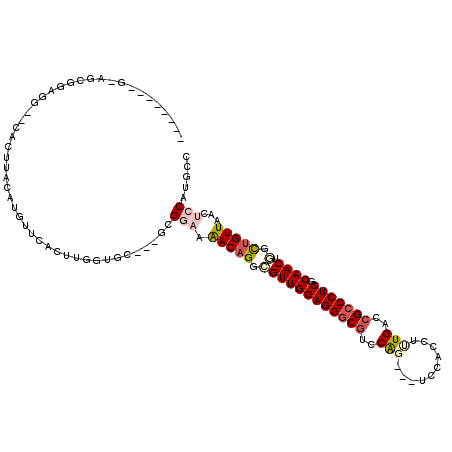
>2R_DroMel_CAF1 16531218 105 + 20766785 ---------CGUAAAUAAUCGAAUAUUGGUGCAGGUACUCGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GC ---------................((((.((((((....(((.((.......))))).((((.(((((((.(((........---.))).)))))))))))))))))...))))---.. ( -40.90) >DroVir_CAF1 23137 113 + 1 AUUAAAUUGUAUACAUACAUGGAU-GGGGUGCAGGUUCUUGGCAUGGAGGUAACAGCCAGUUGCGAGGGCGGUCAAAGAUGGA---CUGGACGCCCUCCAACGCCUGUCUUCCGC---GC .......((((....))))..(.(-((((.((((((...((((.((.......))))))((((.(((((((..((........---.))..))))))))))))))))).))))))---.. ( -41.20) >DroGri_CAF1 17024 103 + 1 ---------UGUAC-UAUAUACAU-GUGGUGCAGGUUCUUGGCAUGGAGUUAACAACUAGUUGCGAGGGCGGUCAAAGAUGGA---CCGGAAGACCUCCAACGCCAGUUUAUCGC---AC ---------(((((-((((....)-))))))))(((..(((((....(((.....))).((((.((((.(((((.......))---))).....)))))))))))))...)))..---.. ( -33.80) >DroWil_CAF1 34071 109 + 1 ----------GUAAUAAAUCU-AGCUCGAUUUAGGUUUUGGGCAUGGAGUUAACAGCCAGUUGUGAGGGCGGCCAAAAAUGGAAUGCUGGACGCCCUCCAACUCCUGUUUUCCGAACAGC ----------...........-.((((((........)))))).(((((..(((((..(((((.(((((((.(((............))).)))))))))))).))))))))))...... ( -38.10) >DroMoj_CAF1 17746 104 + 1 ---------UGUAUAUACAUGUAU-GAGCUGCAGGUUCUUGGCAUGGAGCUAACAGCUAGUUGUGAGGGCGGUCAGAGGUGGA---CCGGACGCCCUCCAACACCUGUUUAUCGC---GC ---------.(((((.(((.((..-.(((((..((((((......))))))..))))).((((.(((((((.((...((....---)).))))))))))))))).))).))).))---.. ( -40.50) >DroAna_CAF1 13536 105 + 1 ---------AGUAAAUAAUUGCCUAUUGGUUCAGGUACUGGGCAUGGAGUUAACAGCCAGUUGCGAGGGUGGCCAAAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GC ---------.((((....))))...((((..(((((....(((.((.......))))).((((.(((((((.(((........---.))).))))))))))))))))....))))---.. ( -37.00) >consensus _________UGUAAAUAAAUGCAU_GUGGUGCAGGUUCUUGGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCAAAAAUGGA___CUGGACGCCCUCCAACGCCUGUUUUCCGA___GC .........................((((.((((((....(((.((.......))))).((((.(((((((.((..............)).)))))))))))))))))...))))..... (-28.43 = -28.74 + 0.31)
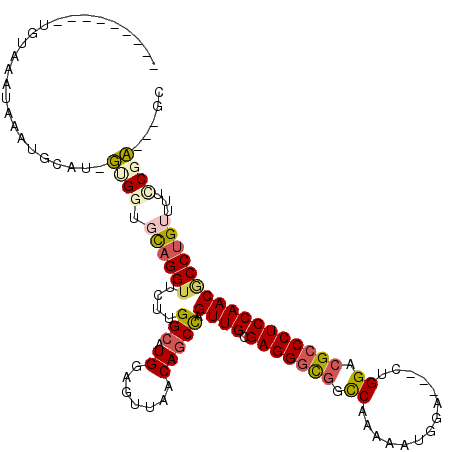
| Location | 16,531,218 – 16,531,323 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.08 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -19.42 |
| Energy contribution | -20.48 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16531218 105 - 20766785 GC---UCGGAAAACAGGCGUUGGAGGGCGUCCAG---UCCACUUGUGGCCGCCCUCGCAACUGGCUGUUAACUCCAUGCCGAGUACCUGCACCAAUAUUCGAUUAUUUACG--------- ((---..(((.(((((.((((((((((((.(((.---........))).))))))).)))).).)))))...)))..))((((((..........))))))..........--------- ( -36.50) >DroVir_CAF1 23137 113 - 1 GC---GCGGAAGACAGGCGUUGGAGGGCGUCCAG---UCCAUCUUUGACCGCCCUCGCAACUGGCUGUUACCUCCAUGCCAAGAACCUGCACCCC-AUCCAUGUAUGUAUACAAUUUAAU ((---(.(((.(((((.((((((((((((..(((---.......)))..))))))).)))).).)))))...))).))).....((.((((....-.....)))).))............ ( -34.90) >DroGri_CAF1 17024 103 - 1 GU---GCGAUAAACUGGCGUUGGAGGUCUUCCGG---UCCAUCUUUGACCGCCCUCGCAACUAGUUGUUAACUCCAUGCCAAGAACCUGCACCAC-AUGUAUAUA-GUACA--------- ((---(((......((((((.((((......(((---((.......))))).....((((....))))...))))))))))......)))))...-.(((((...-)))))--------- ( -28.50) >DroWil_CAF1 34071 109 - 1 GCUGUUCGGAAAACAGGAGUUGGAGGGCGUCCAGCAUUCCAUUUUUGGCCGCCCUCACAACUGGCUGUUAACUCCAUGCCCAAAACCUAAAUCGAGCU-AGAUUUAUUAC---------- .((((((((.....(((((((((((((((.((((.(......).)))).))))))).)))))(((((.......)).))).....)))...)))))).-)).........---------- ( -35.70) >DroMoj_CAF1 17746 104 - 1 GC---GCGAUAAACAGGUGUUGGAGGGCGUCCGG---UCCACCUCUGACCGCCCUCACAACUAGCUGUUAGCUCCAUGCCAAGAACCUGCAGCUC-AUACAUGUAUAUACA--------- ((---(((...(((((.((((((((((((..(((---.......)))..))))))).)))).).))))).((.....))........))).))..-...............--------- ( -32.40) >DroAna_CAF1 13536 105 - 1 GC---UCGGAAAACAGGCGUUGGAGGGCGUCCAG---UCCACUUUUGGCCACCCUCGCAACUGGCUGUUAACUCCAUGCCCAGUACCUGAACCAAUAGGCAAUUAUUUACU--------- ..---..(((.(((((.((((((((((.(.((((---.......)))).).))))).)))).).)))))...))).((((.................))))..........--------- ( -31.93) >consensus GC___GCGGAAAACAGGCGUUGGAGGGCGUCCAG___UCCACCUUUGACCGCCCUCGCAACUGGCUGUUAACUCCAUGCCAAGAACCUGCACCAA_AUGCAAUUAUUUACA_________ .......(((.(((((.((((((((((((..(((..........)))..))))))).)))).).)))))...)))............................................. (-19.42 = -20.48 + 1.06)

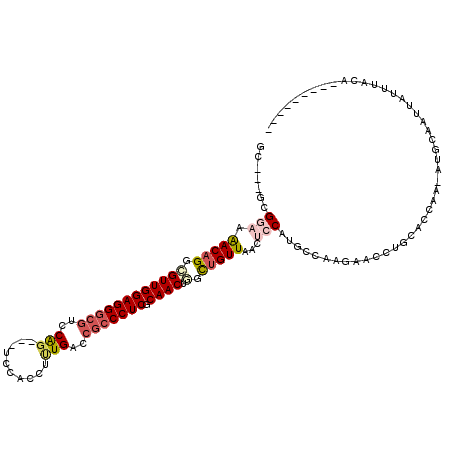
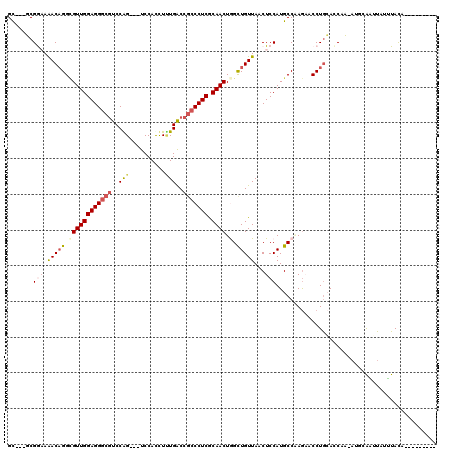
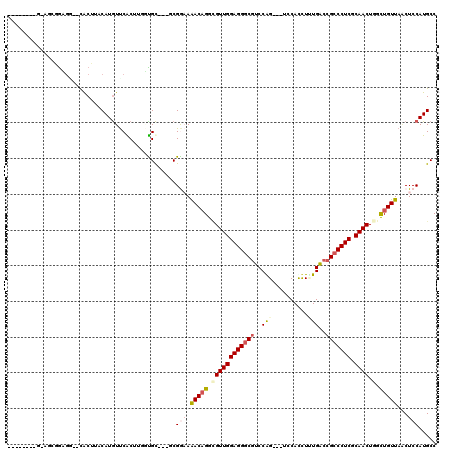
| Location | 16,531,249 – 16,531,356 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -26.13 |
| Energy contribution | -26.19 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16531249 107 + 20766785 GGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGUG--CCACUGACGCACA----- (((((((((..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))))))(.---.(((...)))..)......)))--))...........----- ( -40.50) >DroVir_CAF1 23176 112 + 1 GGCAUGGAGGUAACAGCCAGUUGCGAGGGCGGUCAAAGAUGGA---CUGGACGCCCUCCAACGCCUGUCUUCCGC---GCAAAUAGUGAACAUGUAAGUG--CCUCCGCUGCACCUUAUU .(((((((((((((((...((((.(((((((..((........---.))..)))))))))))..))))....(((---.......)))..........))--)))))).)))........ ( -42.70) >DroGri_CAF1 17053 106 + 1 GGCAUGGAGUUAACAACUAGUUGCGAGGGCGGUCAAAGAUGGA---CCGGAAGACCUCCAACGCCAGUUUAUCGC---ACAAAUAGUGGAUAUGUAAGUGUGCCUUGACUAC-------- ((((((((((.....))).((((.((((.(((((.......))---))).....)))))))).))....((((.(---((.....))))))).......)))))........-------- ( -30.60) >DroWil_CAF1 34100 105 + 1 GGCAUGGAGUUAACAGCCAGUUGUGAGGGCGGCCAAAAAUGGAAUGCUGGACGCCCUCCAACUCCUGUUUUCCGAACAGCACAAAGCGAACAUGUAAGUG--UUGCC------------- (((.(((((..(((((..(((((.(((((((.(((............))).)))))))))))).))))))))))....((.....)).(((((....)))--)))))------------- ( -38.00) >DroYak_CAF1 13975 99 + 1 GGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCACAAGUGGA---CUGGACGCCCUCCAACGCCUGUUUUCCGA---GCGCCAAGCGAACAUGUAAGCA--GCACU------------- (((.(((((..(((((...((((.(((((((.(((........---.))).)))))))))))..)))))))))).---..)))..((..........)).--.....------------- ( -36.40) >DroMoj_CAF1 17776 106 + 1 GGCAUGGAGCUAACAGCUAGUUGUGAGGGCGGUCAGAGGUGGA---CCGGACGCCCUCCAACACCUGUUUAUCGC---GCAUCCAGUGAACAUGUAAGUG--GCUCCGCUCCG------U ((..((((((((((((...((((.(((((((.((...((....---)).)))))))))))))..))))...((((---.......)))).........))--))))))..)).------. ( -43.60) >consensus GGCAUGGAGUUAACAGCCAGUUGCGAGGGCGGCCAAAAAUGGA___CUGGACGCCCUCCAACGCCUGUUUUCCGA___GCACAAAGCGAACAUGUAAGUG__CCUCCGCU_C________ .(((((.....(((((...((((.(((((((.((..............)).)))))))))))..))))).........((.....))...)))))......................... (-26.13 = -26.19 + 0.06)
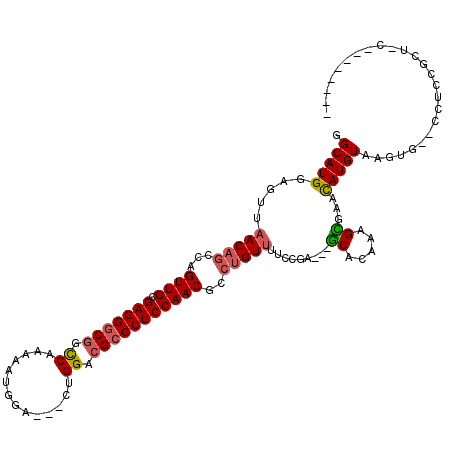
| Location | 16,531,249 – 16,531,356 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.06 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -20.26 |
| Energy contribution | -21.35 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
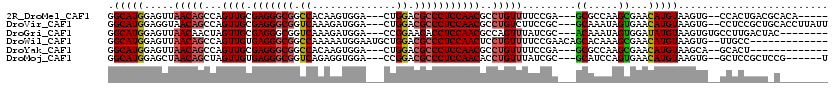
>2R_DroMel_CAF1 16531249 107 - 20766785 -----UGUGCGUCAGUGG--CACUUACAUGUUCGCUUGGCGC---UCGGAAAACAGGCGUUGGAGGGCGUCCAG---UCCACUUGUGGCCGCCCUCGCAACUGGCUGUUAACUCCAUGCC -----.(.((((((((((--.((......)))))).))))))---.)(((.(((((.((((((((((((.(((.---........))).))))))).)))).).)))))...)))..... ( -45.90) >DroVir_CAF1 23176 112 - 1 AAUAAGGUGCAGCGGAGG--CACUUACAUGUUCACUAUUUGC---GCGGAAGACAGGCGUUGGAGGGCGUCCAG---UCCAUCUUUGACCGCCCUCGCAACUGGCUGUUACCUCCAUGCC ........(((..(((((--..(((...(((.((.....)).---))).)))((((.((((((((((((..(((---.......)))..))))))).)))).).))))..))))).))). ( -41.10) >DroGri_CAF1 17053 106 - 1 --------GUAGUCAAGGCACACUUACAUAUCCACUAUUUGU---GCGAUAAACUGGCGUUGGAGGUCUUCCGG---UCCAUCUUUGACCGCCCUCGCAACUAGUUGUUAACUCCAUGCC --------........((((........(((((((.....))---).))))...(((.((((((((.....(((---((.......))))).))))((((....)))))))).))))))) ( -27.90) >DroWil_CAF1 34100 105 - 1 -------------GGCAA--CACUUACAUGUUCGCUUUGUGCUGUUCGGAAAACAGGAGUUGGAGGGCGUCCAGCAUUCCAUUUUUGGCCGCCCUCACAACUGGCUGUUAACUCCAUGCC -------------((((.--.............((.....)).....(((.(((((.((((((((((((.((((.(......).)))).))))))).)))))..)))))...))).)))) ( -37.30) >DroYak_CAF1 13975 99 - 1 -------------AGUGC--UGCUUACAUGUUCGCUUGGCGC---UCGGAAAACAGGCGUUGGAGGGCGUCCAG---UCCACUUGUGGCCGCCCUCGCAACUGGCUGUUAACUCCAUGCC -------------(((((--(((..........))..)))))---).(((.(((((.((((((((((((.(((.---........))).))))))).)))).).)))))...)))..... ( -38.10) >DroMoj_CAF1 17776 106 - 1 A------CGGAGCGGAGC--CACUUACAUGUUCACUGGAUGC---GCGAUAAACAGGUGUUGGAGGGCGUCCGG---UCCACCUCUGACCGCCCUCACAACUAGCUGUUAGCUCCAUGCC .------.(((((.((((--.........)))).........---......(((((.((((((((((((..(((---.......)))..))))))).)))).).))))).)))))..... ( -39.90) >consensus ________G_AGCGGAGG__CACUUACAUGUUCACUUGGUGC___GCGGAAAACAGGCGUUGGAGGGCGUCCAG___UCCACCUUUGACCGCCCUCGCAACUGGCUGUUAACUCCAUGCC ...............................................(((.(((((.((((((((((((..(((..........)))..))))))).)))).).)))))...)))..... (-20.26 = -21.35 + 1.09)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:12 2006