| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,519,018 – 16,519,128 |
| Length | 110 |
| Max. P | 0.653904 |

| Location | 16,519,018 – 16,519,128 |
|---|---|
| Length | 110 |
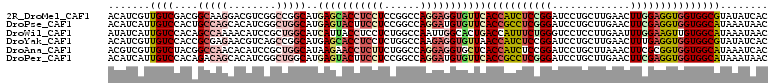
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.58 |
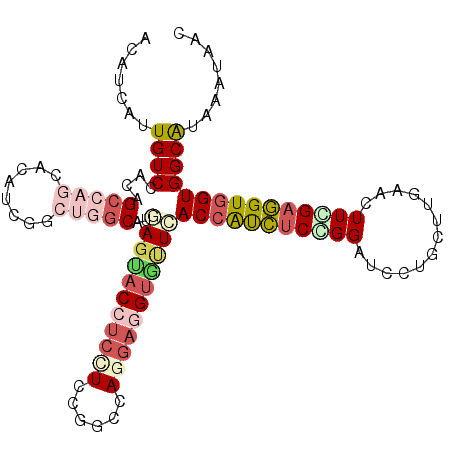
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -24.72 |
| Energy contribution | -26.55 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16519018 110 + 20766785 ACAUCGUUGUCGACGGCAAGGACGUCGGCCGGCAUGAGCACCUCCUCCGGCCAGGAGGUGUUCACCAUCUCCGGAUCCUGCUUGAACUUGGAGGUGGUGGCGUAUAUCAC ........(((((((.......)))))))((.(..(((((((((((......)))))))))))(((((((((((.((......)).).))))))))))).))........ ( -47.80) >DroPse_CAF1 1618 110 + 1 ACAUCAUUGUCCACUGCCAGCACAUCGGCUGGCAUGAGUACUUCCUCCGGCCAGGAUGUGUUCACCGCCUCGGGAUCCUGCUUGAACUUCGAGGUGGUGGCAUAAAUAAC (((((.(((.((..(((((((......))))))).(((......))).)).))))))))((.((((((((((((.((......)).).)))))))))))))......... ( -42.90) >DroWil_CAF1 1297 110 + 1 AUAUCAUUGUCCACAGCCAAAACAUCCGCUGGCAUCAUUACCUCCUCUGGCCAAUUGGCACUGACCAUUUCUGGGUCCUCCUUGAAUUUGGAAGUUGUGGCAUAAAUAAC ......((((((((((((((.....(((..((...........))..)))....)))))(((..(((.(((.(((....))).)))..))).)))))))).))))..... ( -25.70) >DroYak_CAF1 1567 110 + 1 ACAUCGUUGUCCACCGCGAGAACGUCAGCCGGCAUGAGCACCUCCUCUGGCCAAGAGGUGUUAACCAUCUCCGGAUCCUGCUUGAACUUUGAGGUGGUGGCGUAUAUCAC ....((((.((......)).))))...(((......((((((((..........)))))))).((((((((.((.((......)).))..)))))))))))......... ( -31.80) >DroAna_CAF1 1569 110 + 1 ACGUCGUUGUCUACGGCCAACACAUCCGCUGGCAUAAGAACCUCUUCUGGCCAGGAGGUGCUCACCAUCUCCGGAUCCUGCUUAAACUUCGCGGUGGUGGCAUAAAUCAC ..((.((((.(....).)))))).....(((((...((((....)))).)))))((.((((.(((((((..((((............)))).)))))))))))...)).. ( -35.30) >DroPer_CAF1 1615 110 + 1 ACAUCAUUGUCCACAGACAGCACAUCGGCUGGCAUGAGUACUUCCUCCGGCCAGGAUGUGUUCACCGCCUCGGGAUCCUGCUUGAACUUCGAGGUGGUGGCAUAAAUAAC .......((((....))))(((((((..(((((..(((......)))..))))))))))))(((((((((((((.((......)).).)))))))))))).......... ( -41.70) >consensus ACAUCAUUGUCCACAGCCAGCACAUCGGCUGGCAUGAGUACCUCCUCCGGCCAGGAGGUGUUCACCAUCUCCGGAUCCUGCUUGAACUUCGAGGUGGUGGCAUAAAUAAC .......((((....(((((........)))))..(((((((((((......)))))))))))(((((((((((.............)))))))))))))))........ (-24.72 = -26.55 + 1.84)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:08 2006