
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,508,882 – 16,508,989 |
| Length | 107 |
| Max. P | 0.860036 |

| Location | 16,508,882 – 16,508,989 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -21.21 |
| Energy contribution | -22.47 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
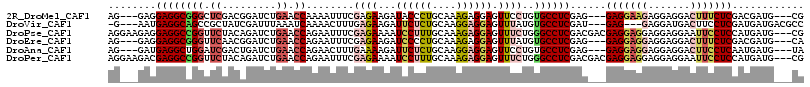
>2R_DroMel_CAF1 16508882 107 + 20766785 AG---GAGGAGGCGGGCUCGACGGAUCUGAACCAAAAUUUCGAGAAGAUACCCUGCAAAGAGGAGUUCCUGUGCCUCGAG---GAGGAAGAGGAGGACUUUCUCGACGAUG---CG .(---(.(((..((.......))..)))...))......((((((((...((((....)).))..(((((.(.((((...---)))).).)))))...)))))))).....---.. ( -27.60) >DroVir_CAF1 11766 106 + 1 -G---AAUGAGGCAGCCGCUAUCGAUUUAAAUCAAAACUUUGAGAAGAUUCUCUGCAAGGAGGAGUUUAUGUGCCUCGAU---GAG---GAGGAUGACUUCCUCGAUGAUGACGCC -.---..(((((((.......(((((....)))....((((((((......))).)))))..)).......))))))).(---(((---((((....))))))))........... ( -27.74) >DroPse_CAF1 11767 113 + 1 AGGAAGAGGAGGCCGGUUCUACAGAUCUGAACCAGAAUUUCGAGAAAAUCCUUUGCAAAGAGGAGUUUCUGGGCCUCGACGACGAGGAGGAGGAGGAAUUCCUCCAUGAUG---CG ........(((((((((((.........))))).........(((((.((((((....)))))).))))).))))))(.((.((.(((((((......))))))).)).))---). ( -42.20) >DroEre_CAF1 6070 107 + 1 AG---GAGGAGGCGGGUUCAACGGAUCUGAACCAGAAUUUCGAGAAGAUCCCCUGCAAAGAGGAGUUUAUGUGCCUCGAG---GAGGAGGAGGAGGACUUUCUCGACGAUG---CA ..---......(((((((((.......))))))......((((((((.(((((((((.(((....))).))).((((...---...))))))).))).))))))))...))---). ( -33.10) >DroAna_CAF1 5771 107 + 1 AG---GAUGAGGCUGGAUCGACUGAUCUGAACCAGAACUUUGAAAAGAUUCUCUGCAAGGAGGAGUUCCUGUGCCUCGAG---GAGGAGGAGGAGGACUUCCUCAAUGAUG---UA ..---..((((((.(((.(..((..((((...)))).(((((...((.....)).)))))))..).)))...))))))..---...(((((((....))))))).......---.. ( -32.70) >DroPer_CAF1 11704 113 + 1 AGGAAGACGAGGCCGGUUCUACAGAUCUGAACCAGAAUUUCGAGAAAAUCCUUUGCAAAGAGGAGUUUCUGGGCCUCGACGACGAGGAGGAGGAGGAAUUCCUCCAUGAUG---CG .......((((((((((((.........))))).........(((((.((((((....)))))).))))).))))))).......(((((((......)))))))......---.. ( -44.20) >consensus AG___GAGGAGGCCGGUUCUACAGAUCUGAACCAGAAUUUCGAGAAGAUCCUCUGCAAAGAGGAGUUUCUGUGCCUCGAG___GAGGAGGAGGAGGACUUCCUCGAUGAUG___CG ........((((((((.((.........)).))........((((...((((((....)))))).))))..))))))......(((((((.......)))))))............ (-21.21 = -22.47 + 1.25)

| Location | 16,508,882 – 16,508,989 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.45 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.860036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
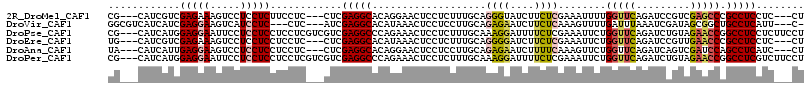
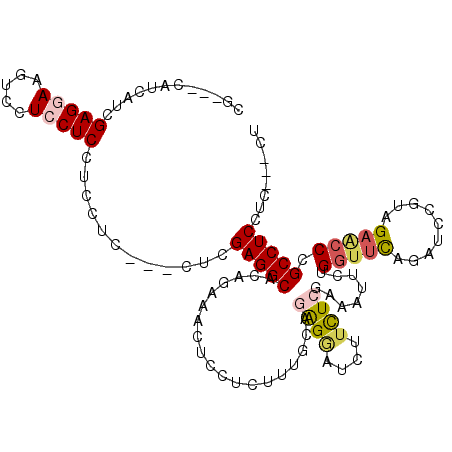
>2R_DroMel_CAF1 16508882 107 - 20766785 CG---CAUCGUCGAGAAAGUCCUCCUCUUCCUC---CUCGAGGCACAGGAACUCCUCUUUGCAGGGUAUCUUCUCGAAAUUUUGGUUCAGAUCCGUCGAGCCCGCCUCCUC---CU .(---(....(((((((.....((((...((((---...))))...))))((.(((......)))))...)))))))......(((((.((....))))))).))......---.. ( -26.30) >DroVir_CAF1 11766 106 - 1 GGCGUCAUCAUCGAGGAAGUCAUCCUC---CUC---AUCGAGGCACAUAAACUCCUCCUUGCAGAGAAUCUUCUCAAAGUUUUGAUUUAAAUCGAUAGCGGCUGCCUCAUU---C- ((((((...(((((..((((((.((((---...---...))))....................((((....)))).......))))))...)))))...))).))).....---.- ( -25.00) >DroPse_CAF1 11767 113 - 1 CG---CAUCAUGGAGGAAUUCCUCCUCCUCCUCGUCGUCGAGGCCCAGAAACUCCUCUUUGCAAAGGAUUUUCUCGAAAUUCUGGUUCAGAUCUGUAGAACCGGCCUCCUCUUCCU ..---......((((((.....))))))........(..((((((.(((((.((((........)))).))))).........(((((.........)))))))))))..)..... ( -34.70) >DroEre_CAF1 6070 107 - 1 UG---CAUCGUCGAGAAAGUCCUCCUCCUCCUC---CUCGAGGCACAUAAACUCCUCUUUGCAGGGGAUCUUCUCGAAAUUCUGGUUCAGAUCCGUUGAACCCGCCUCCUC---CU .(---(....(((((((..(((.(((.(((...---...)))(((..............)))))))))..)))))))......(((((((.....))))))).))......---.. ( -29.54) >DroAna_CAF1 5771 107 - 1 UA---CAUCAUUGAGGAAGUCCUCCUCCUCCUC---CUCGAGGCACAGGAACUCCUCCUUGCAGAGAAUCUUUUCAAAGUUCUGGUUCAGAUCAGUCGAUCCAGCCUCAUC---CU ..---...((..(((((..((((...((((...---...))))...))))..)))))..))...((((.(((....)))))))((((..((((....)))).)))).....---.. ( -27.30) >DroPer_CAF1 11704 113 - 1 CG---CAUCAUGGAGGAAUUCCUCCUCCUCCUCGUCGUCGAGGCCCAGAAACUCCUCUUUGCAAAGGAUUUUCUCGAAAUUCUGGUUCAGAUCUGUAGAACCGGCCUCGUCUUCCU ..---......((((((.....))))))........(.(((((((.(((((.((((........)))).))))).........(((((.........)))))))))))).)..... ( -37.70) >consensus CG___CAUCAUCGAGGAAGUCCUCCUCCUCCUC___CUCGAGGCACAGAAACUCCUCUUUGCAGAGGAUCUUCUCGAAAUUCUGGUUCAGAUCCGUAGAACCCGCCUCCUC___CU ............(((((.....)))))............(((((...................((((....))))........(((((.........))))).)))))........ (-16.61 = -17.12 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:36:02 2006