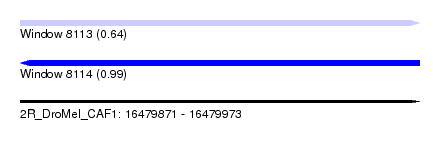
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,479,871 – 16,479,973 |
| Length | 102 |
| Max. P | 0.991707 |

| Location | 16,479,871 – 16,479,973 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 55.70 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -10.93 |
| Energy contribution | -13.79 |
| Covariance contribution | 2.87 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.32 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16479871 102 + 20766785 UCAUUUCCGCAACACACACGCAGUUGCAGUUGCAUACGCACU-UGCCACAGUUGCGGCAGUUGCUGUUGCAGCAGUUGGUGCAGUUGCAGCUGCUGCGAUUGC ........(((((.........)))))(((.((....)))))-.....((((((((((((((((..(((((.(....).)))))..)))))))))))))))). ( -43.10) >DroSec_CAF1 155123 79 + 1 UCAUUUCCGCAGCACACACGCAGUUGCAGUUGCAGUUGCACUUUGCCACUGUUGCGGCAGUU------------------------GCAGCUGCUGCGGNNNN ......((((((((.....(((..(((.((.(((((.((.....)).))))).)).)))..)------------------------))...)))))))).... ( -33.80) >DroAna_CAF1 153127 85 + 1 UCAUUUCCGUGGCACACCCGCAACAUCAGUGGCACU-------UUCCACUUCU--GACAGUGGCAGUUGCAGUGGCAC-GGCAGUU--------UGCGGUGGC ..(((.(((((.(((...(((.......)))(((.(-------(.(((((...--...))))).)).))).))).)))-)).))).--------......... ( -25.00) >consensus UCAUUUCCGCAGCACACACGCAGUUGCAGUUGCACU_GCACU_UGCCACUGUUGCGGCAGUUGC_GUUGCAG__G_____GCAGUUGCAGCUGCUGCGGU_GC ........(((((...............)))))..................(((((((((((....(((((........)))))....))))))))))).... (-10.93 = -13.79 + 2.87)

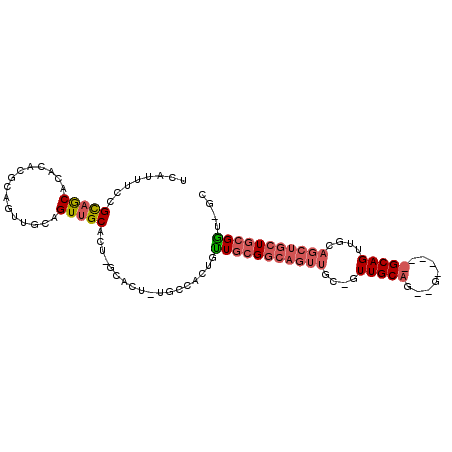
| Location | 16,479,871 – 16,479,973 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 55.70 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -13.23 |
| Energy contribution | -15.23 |
| Covariance contribution | 2.01 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16479871 102 - 20766785 GCAAUCGCAGCAGCUGCAACUGCACCAACUGCUGCAACAGCAACUGCCGCAACUGUGGCA-AGUGCGUAUGCAACUGCAACUGCGUGUGUGUUGCGGAAAUGA (((...(((((((.(((....)))....)))))))....(((..((((((....))))))-..)))...)))..(((((((.........)))))))...... ( -42.30) >DroSec_CAF1 155123 79 - 1 NNNNCCGCAGCAGCUGC------------------------AACUGCCGCAACAGUGGCAAAGUGCAACUGCAACUGCAACUGCGUGUGUGCUGCGGAAAUGA ....(((((((((.(((------------------------(..((((((....))))))...)))).))(((..(((....)))..))))))))))...... ( -33.80) >DroAna_CAF1 153127 85 - 1 GCCACCGCA--------AACUGCC-GUGCCACUGCAACUGCCACUGUC--AGAAGUGGAA-------AGUGCCACUGAUGUUGCGGGUGUGCCACGGAAAUGA (((((((((--------(....(.-(((.((((.......(((((...--...)))))..-------)))).))).)...)))).)))).))........... ( -27.30) >consensus GC_ACCGCAGCAGCUGCAACUGC_____C__CUGCAAC_GCAACUGCCGCAACAGUGGCA_AGUGC_AAUGCAACUGCAACUGCGUGUGUGCUGCGGAAAUGA ....((((((((................................((((((....))))))..........(((...((....)).))).))))))))...... (-13.23 = -15.23 + 2.01)

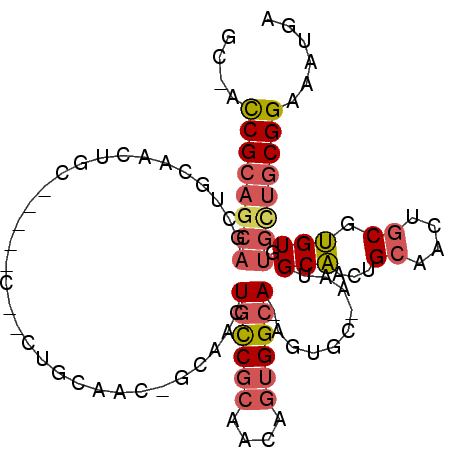
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:53 2006