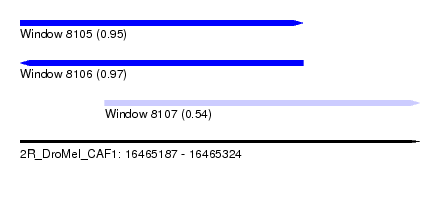
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,465,187 – 16,465,324 |
| Length | 137 |
| Max. P | 0.970744 |

| Location | 16,465,187 – 16,465,284 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -20.60 |
| Consensus MFE | -11.97 |
| Energy contribution | -12.22 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.949965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
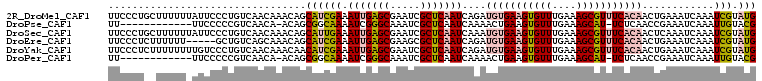
>2R_DroMel_CAF1 16465187 97 + 20766785 U--UUUUCAC-UUUUUGUGCCA-GGCCAUACUUUUCCCUGCUUUUUUAUUCCCUGUCAACAAACAGCAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGA .--.......-..(((((..((-((..(((................)))..))))...)))))..(((((...((((((((...))))))))..))))).. ( -20.99) >DroSec_CAF1 140470 96 + 1 UCAUUU----UUUUUUUUGCCA-GGCGAUACUUUUCCCUGCUUUUUUAUUCCCUGUCAACAAACAGCAUUGAAAUUGAGCGAAUCGCUCAAUCAAAUGUGA ......----..(((.(((.((-((.((((................)))).)))).))).)))..(((((...((((((((...))))))))..))))).. ( -19.39) >DroSim_CAF1 137215 100 + 1 UCAUUUUCAUUUUUUUGUGCCA-GGCAAUACUUUUCCCUGCUUUUUUAUUCCCUGUCAACAAACAACAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGA .............(((((..((-((.((((................)))).))))...)))))..(((((...((((((((...))))))))..))))).. ( -22.39) >DroEre_CAF1 141740 86 + 1 --------AU-UUUCUGUACCA-GGCGAUACUUUUCCCUCUUUUUU-----GCUGUCAGCAAACAGCAUCGAAAUUGAGCGAAGCGCUCAAUCAGAUGUGA --------..-...((((...(-((.((......))))).....((-----((.....))))))))((((...((((((((...))))))))..))))... ( -21.90) >DroYak_CAF1 142263 95 + 1 UU----GGAU-UUUCUGUAGCA-GGCGAUACUUUUCCCUCUUUUUUUUGUCCCUGUCAACAAACAACAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGA ..----((((-.......((.(-((.((......))))))).......)))).............(((((...((((((((...))))))))..))))).. ( -23.84) >DroPer_CAF1 174794 76 + 1 ------------UUUUGUGCGAUUGCGAUACUUUU------------UUCCCCCGUCAACA-ACAGCGGCAAAAUCGGGCAAAUCGCUCAAUCAAAACUGA ------------..(((.((((((((((.......------------.))..((((.....-...)))).........)).)))))).))).......... ( -15.10) >consensus U_______AU_UUUUUGUGCCA_GGCGAUACUUUUCCCUGCUUUUUUAUUCCCUGUCAACAAACAGCAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGA .................................................................(((((...(((((((.....)))))))..))))).. (-11.97 = -12.22 + 0.25)


| Location | 16,465,187 – 16,465,284 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 76.00 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.07 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16465187 97 - 20766785 UCACAUCUGAUUGAGCGAUUCGCUCAAUUUCGAUGCUGUUUGUUGACAGGGAAUAAAAAAGCAGGGAAAAGUAUGGCC-UGGCACAAAAA-GUGAAAA--A (((((((.(((((((((...)))))))))..))).(((((....)))))...........((.(((..(....)..))-).)).......-))))...--. ( -24.40) >DroSec_CAF1 140470 96 - 1 UCACAUUUGAUUGAGCGAUUCGCUCAAUUUCAAUGCUGUUUGUUGACAGGGAAUAAAAAAGCAGGGAAAAGUAUCGCC-UGGCAAAAAAAA----AAAUGA ...((((((((((((((...)))))))))....(.(((((....))))).)..........((((((......)).))-))..........----))))). ( -22.20) >DroSim_CAF1 137215 100 - 1 UCACAUCUGAUUGAGCGAUUCGCUCAAUUUCGAUGUUGUUUGUUGACAGGGAAUAAAAAAGCAGGGAAAAGUAUUGCC-UGGCACAAAAAAAUGAAAAUGA ..(((((.(((((((((...)))))))))..)))))..(((((...((((.((((................)))).))-))..)))))............. ( -24.99) >DroEre_CAF1 141740 86 - 1 UCACAUCUGAUUGAGCGCUUCGCUCAAUUUCGAUGCUGUUUGCUGACAGC-----AAAAAAGAGGGAAAAGUAUCGCC-UGGUACAGAAA-AU-------- ((...((((((((((((...)))))))))....(((((((....))))))-----)....)))..))...(((((...-.))))).....-..-------- ( -24.90) >DroYak_CAF1 142263 95 - 1 UCACAUCUGAUUGAGCGAUUCGCUCAAUUUCGAUGUUGUUUGUUGACAGGGACAAAAAAAAGAGGGAAAAGUAUCGCC-UGCUACAGAAA-AUCC----AA ..(((((.(((((((((...)))))))))..)))))..(((((.(.((((((.....................)).))-))).)))))..-....----.. ( -22.60) >DroPer_CAF1 174794 76 - 1 UCAGUUUUGAUUGAGCGAUUUGCCCGAUUUUGCCGCUGU-UGUUGACGGGGGAA------------AAAAGUAUCGCAAUCGCACAAAA------------ (((((....)))))((((((.((...(((((.((.((((-(...))))).))..------------.)))))...))))))))......------------ ( -22.30) >consensus UCACAUCUGAUUGAGCGAUUCGCUCAAUUUCGAUGCUGUUUGUUGACAGGGAAUAAAAAAGCAGGGAAAAGUAUCGCC_UGGCACAAAAA_AU_______A ...((((.(((((((((...)))))))))..))))(((((....)))))..........................((....)).................. (-15.28 = -15.07 + -0.22)

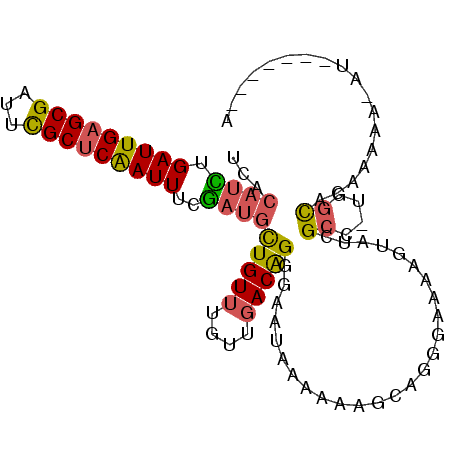
| Location | 16,465,216 – 16,465,324 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -14.07 |
| Energy contribution | -14.60 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.537081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
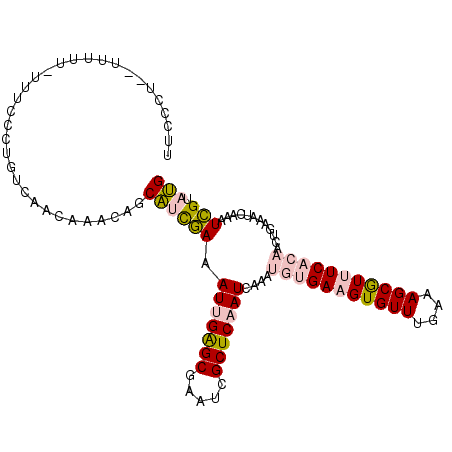
>2R_DroMel_CAF1 16465216 108 + 20766785 UUCCCUGCUUUUUUAUUCCCUGUCAACAAACAGCAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGAAGUGUUUGAAAGCGUUUCACAACUGAAAUCAAAUCGUAUG ...................((((......))))((((((..(((((((...)))))))((((.(((((((((((....))))))))))).)))).......))).))) ( -27.00) >DroPse_CAF1 173877 94 + 1 UU------------UUCCCCCGUCAACA-ACAGCGGCAAAAUCGGGCAAAUCGCUCAAUCAAAACUGAAGUGUUUGAAAGCAU-UCUCAACCGAAAUCAAAUUGUACG ..------------......((((((..-....(((.......((((.....)))).........(((((((((....)))))-).))).)))........))).))) ( -13.84) >DroSec_CAF1 140498 108 + 1 UUCCCUGCUUUUUUAUUCCCUGUCAACAAACAGCAUUGAAAUUGAGCGAAUCGCUCAAUCAAAUGUGAAGUGUUUGAAAGCGUUUCACAACUCAAAUCAAAUCGUAUG .....(((...........((((......))))..((((.((((((((...))))))))....(((((((((((....))))))))))).......))))...))).. ( -21.90) >DroEre_CAF1 141763 103 + 1 UUCCCUCUUUUUU-----GCUGUCAGCAAACAGCAUCGAAAUUGAGCGAAGCGCUCAAUCAGAUGUGAAGUGUUUGAAAGCGUUUCACAACUGAAAUCAAAUCGUAUG ............(-----(((((......)))))).(((..(((((((...)))))))((((.(((((((((((....))))))))))).)))).......))).... ( -30.80) >DroYak_CAF1 142290 108 + 1 UUCCCUCUUUUUUUUGUCCCUGUCAACAAACAACAUCGAAAUUGAGCGAAUCGCUCAAUCAGAUGUGAAGUGUUUGAAAGCGUUUCACAACUGAAAUCAAAUCGUAUG ............(((((........)))))...((((((..(((((((...)))))))((((.(((((((((((....))))))))))).)))).......))).))) ( -26.30) >DroPer_CAF1 174815 94 + 1 UU------------UUCCCCCGUCAACA-ACAGCGGCAAAAUCGGGCAAAUCGCUCAAUCAAAACUGAAGUGUUUGAAAGCAU-UCUCAACCGAAAUCAAAUUGUACG ..------------......((((((..-....(((.......((((.....)))).........(((((((((....)))))-).))).)))........))).))) ( -13.84) >consensus UUCCCU__UUUUU_UUUCCCUGUCAACAAACAGCAUCGAAAUUGAGCGAAUCGCUCAAUCAAAUGUGAAGUGUUUGAAAGCGUUUCACAACUGAAAUCAAAUCGUAUG .................................((((((.(((((((.....)))))))....(((((((((((....)))))))))))............))).))) (-14.07 = -14.60 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:46 2006