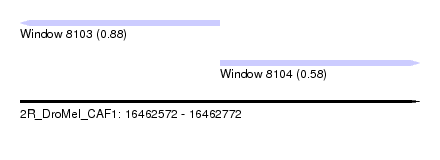
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,462,572 – 16,462,772 |
| Length | 200 |
| Max. P | 0.877404 |

| Location | 16,462,572 – 16,462,672 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 69.54 |
| Mean single sequence MFE | -42.48 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.29 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877404 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16462572 100 - 20766785 AA---GCUAGUGCCUCAUCC---CACACCGCCGCAUGC---CACGCCCCCAGCGGGCGGCAACGGAGGCGGCGGACUGCUAACGGGCGGACUGAU------CUCGACGGCCGCCC .(---((..(((........---))).(((((((...(---(..((((.....))))(....)))..)))))))...)))...((((((.(((..------.....))))))))) ( -47.20) >DroPse_CAF1 171266 94 - 1 AAACCGCUCGUGCCUCAUCC------------CCACGC---CACGCCACCCUCGGGU------GGAGGCACCACCCUUAUCUCGGGUGGCCUGAUGCUCACUUCGACGGCAGCCC ...(((.(((.((.(((...------------....((---(...(((((....)))------)).))).((((((.......))))))..))).))......))))))...... ( -34.80) >DroSec_CAF1 137899 100 - 1 AA---GCUAGUGCCUCAUCC---CACGCCGCCCCAUGC---CACGCCCCCAGAGGGCGGCAGCGGAGGCAGUGGACUGCUCACGGGCGGACUGAU------CUCGACGGCCGCCC .(---((((.((((((....---...(((((((..((.---........))..)))))))....)))))).))).))......((((((.(((..------.....))))))))) ( -45.10) >DroYak_CAF1 139618 100 - 1 AA---GCUAGUGCCUCAUCC---CACGCCGCCCCAUGC---CACGCCCCCAGCGGGCGGCAGCGGGGGCGACGGUCUGCUCACGGGCGGACUGAU------CUCGACGGCCGCCC ..---((..(((........---)))((((((((.(((---(..((((.....))))))))...)))))(((((((((((....))))))))).)------).....))).)).. ( -48.70) >DroAna_CAF1 135122 106 - 1 AA---ACUGGUGGCCCAUCAGCAUCCCCCGCCACAUGCGGUCUCACCCUCCGGGGGC------GGUGCCACCACCCUGAUCUCGGGCGGCCUGAUGCUCACCUCUACGGCCGCCC ..---..(((((((((...........((((.....)))).....(((....)))..------)).)))))))..........((((((((.((........))...)))))))) ( -44.30) >DroPer_CAF1 172201 94 - 1 AAACCGCUCGUGCCUCAUCC------------CCACGC---CACGCCACCCUCGGGU------GGAGGCACCACCCUUAUCUCGGGUGGCCUGAUGCUCACUUCGACGGCAGCCC ...(((.(((.((.(((...------------....((---(...(((((....)))------)).))).((((((.......))))))..))).))......))))))...... ( -34.80) >consensus AA___GCUAGUGCCUCAUCC___CAC_CCGCCCCAUGC___CACGCCCCCAGCGGGC______GGAGGCACCACCCUGAUCACGGGCGGACUGAU______CUCGACGGCCGCCC ..........((((((....................(......)((((.....)))).......)))))).............((((((.(((.............))))))))) (-22.28 = -22.29 + 0.00)

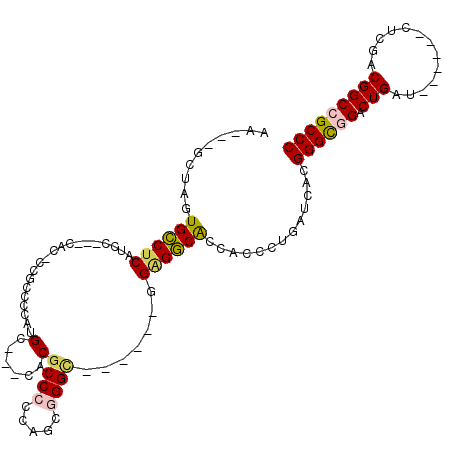
| Location | 16,462,672 – 16,462,772 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.87 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16462672 100 + 20766785 GGGACUCUCGCGCGGACUGCUGUUGCUGCACGGCGGGCUAACUUGGCUCAUCGCAGAGAGUCCUGCGUGCAGCGAUAGAUUCUGCGGAUAGCCGGCCGAU--- .((.((.((.((((((...(((((((((((((..(((((.....)))))..)((((......))))))))))))))))..)))))))).)))).......--- ( -48.90) >DroGri_CAF1 163059 94 + 1 GGGACUUUCGCGAGGGCUGCUGUUGCUGCACGGCGGACUGACCUGCGCC---GACG------AUGGAUGCAGCGAGAGAUUCUGCGGGUAGCCGGCCGAGGCA .((.(((((((.((((...((.(((((((((((((.(......).))))---).(.------...).)))))))).)).))))))))).)))).((....)). ( -35.00) >DroSec_CAF1 137999 100 + 1 GGGACUCUCGCGCGGACUGCUGUUGCUGCACGGCGGGCUCACCUGGCUCAUUGCAGAGAGACCAGCGUGCAGCGAUAGAUUCUGCGGGUAGCCGGCCGAG--- .((.((..(.((((((...(((((((((((((.(.(((((..(((.(.....)))).))).)).))))))))))))))..)))))).).)))).......--- ( -45.60) >DroEre_CAF1 139256 100 + 1 GGGACUCUCGCGCGGACUGCUGUUGCUGCAGGGCGGGCUCACCUGGCUCAUCGCGGAGAGUCCGGCGUGCAGCGAAAGGUUCUGCGGAUAGCCGGCGGAU--- .((.((.((.((((((((....((((((((.(.(((((((.((..((.....)))).))))))).).))))))))..)).)))))))).)))).......--- ( -49.30) >DroYak_CAF1 139718 100 + 1 GGGACUCUCGCGCGGACUGCUGUUGCUGCAGGGCGGGCUCACCUGGCUCAUCGCCGAAAGAUUGGCGUGCAGCGACAGGUUCUGCGGAUAGCCGGCGGAA--- .((.((.((.((((((...((((((((((((((((((....))).))))..((((((....)))))))))))))))))..)))))))).)))).......--- ( -48.30) >DroMoj_CAF1 226204 100 + 1 GGGGCUCUCCCGGGGACUGCUGUUGCUGCAGGGCGGGCUCACCUGCGCC---GACGACGACGACGAAUGCAGCGACAAGUUCUGCGGAUAGCCAACGGAGACA ..((((.(((..((((((..(((((((((((((((((....))))).))---..((....)).....))))))))))))))))..))).))))...(....). ( -45.50) >consensus GGGACUCUCGCGCGGACUGCUGUUGCUGCACGGCGGGCUCACCUGGCUCAUCGCCGAGAGACCGGCGUGCAGCGACAGAUUCUGCGGAUAGCCGGCCGAG___ .((.((.((.((((((...(((((((((((...((((....)))).(((......))).........)))))))))))..)))))))).)))).......... (-29.98 = -30.87 + 0.89)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:44 2006