| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
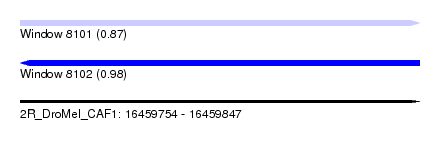
| Location | 16,459,754 – 16,459,847 |
| Length | 93 |
| Max. P | 0.980166 |

| Location | 16,459,754 – 16,459,847 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 74.73 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -14.93 |
| Energy contribution | -15.93 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
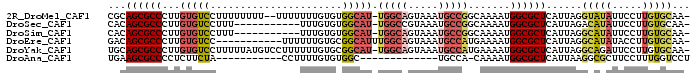
>2R_DroMel_CAF1 16459754 93 + 20766785 CGCAGCGCCCUUGUGUCCUUUUUUUU--UUUUUUUGUGUGGCAU-UGGCAGUAAAUGCCGGCAAAAUGGCGCUCAUUAGGUAUAUUCCUUGUGCAA- .((((((((.((.((((.........--......((.....)).-.((((.....)))))))).)).)))))).((.(((......))).))))..- ( -24.40) >DroSec_CAF1 135070 84 + 1 CACAGCGCCCUUGUGUCCUUU-----------UUUGUGUGGCAU-UGGCCGUAAAUGCCGGCAAAAUGGCGCUCAUUAGACAUAUUCCUUGUGCAA- ....((((...((((((....-----------..((.(((.(((-(.((((.......))))..)))).))).))...))))))......))))..- ( -24.70) >DroSim_CAF1 131878 84 + 1 CACAGCGCCCUUGUGUCCUUU-----------UUUGUGUGGCAU-UGGCAGUAAAUGCCGGCAAAAUGGCGCUCAUUAGGCAUAUUCCUUGUGCAA- ...((((((.((.(((((...-----------...)...(((((-(.......)))))))))).)).))))))......(((((.....)))))..- ( -25.80) >DroEre_CAF1 136168 85 + 1 GACAGCGCCCUUGUGUCC-----------UUUUUUGUGCGGCAUUUGGCAGUAAAUGCCAUGAAAAUGGCGCUCAUUAGGCAUAUACCUUGUGCAA- ....((((...((((.((-----------(....((.(((.(((((((((.....)))))....)))).))).))..)))))))......))))..- ( -28.00) >DroYak_CAF1 136584 95 + 1 UGCAGCGCCCUUGUGUCCUUUUUAUGUCCUUUUUUGUGCGGCAU-UGGCAGUAAAUGCCAUGAAAAUGGCGCUCAUUAGGCAGAUUCCUUGUGCAA- ((((((((....))))........((.(((....((.(((.(((-(((((.....))))).....))).))).))..))))).........)))).- ( -24.90) >DroAna_CAF1 131732 72 + 1 UGAAGCGCCCCUCUUCUA-----------CCUUUUGUGUGGC-------------UGCCA-CAAAAUGGCGCUCAUUAAGGCGCUUCCUUUGGUCCU .((((((((......(((-----------((....).)))).-------------.((((-.....)))).........)))))))).......... ( -21.80) >consensus CACAGCGCCCUUGUGUCCUUU________UUUUUUGUGUGGCAU_UGGCAGUAAAUGCCAGCAAAAUGGCGCUCAUUAGGCAUAUUCCUUGUGCAA_ ...((((((...(((((......................))))).(((((.....))))).......))))))......(((((.....)))))... (-14.93 = -15.93 + 1.01)
| Location | 16,459,754 – 16,459,847 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Mean single sequence MFE | -23.62 |
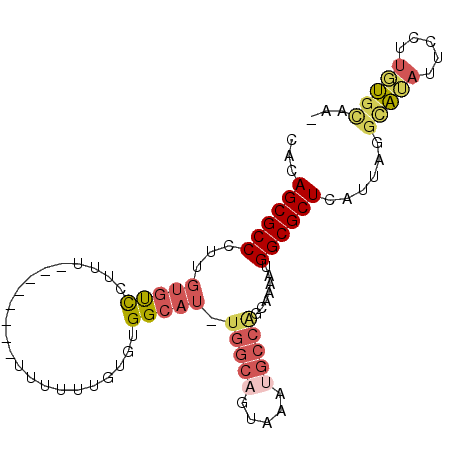
| Consensus MFE | -16.43 |
| Energy contribution | -16.08 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
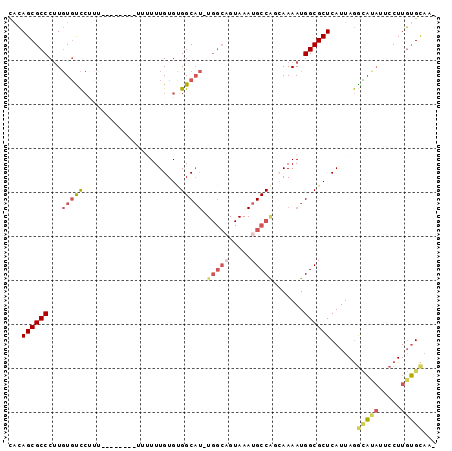
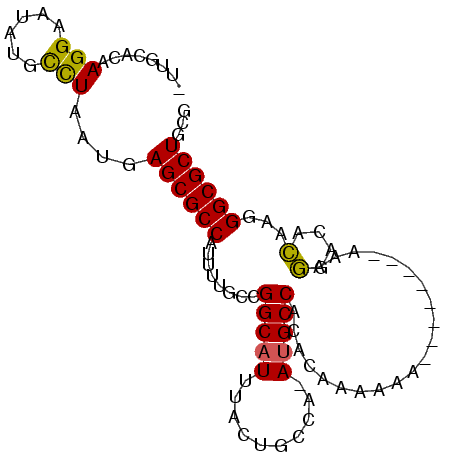
>2R_DroMel_CAF1 16459754 93 - 20766785 -UUGCACAAGGAAUAUACCUAAUGAGCGCCAUUUUGCCGGCAUUUACUGCCA-AUGCCACACAAAAAAA--AAAAAAAAGGACACAAGGGCGCUGCG -..((...(((......)))....((((((.((.((((((((.....)))).-.((.....))......--........)).)).)).)))))))). ( -23.10) >DroSec_CAF1 135070 84 - 1 -UUGCACAAGGAAUAUGUCUAAUGAGCGCCAUUUUGCCGGCAUUUACGGCCA-AUGCCACACAAA-----------AAAGGACACAAGGGCGCUGUG -...(((..(((.....)))....((((((.(((((..((((((.......)-)))))...))))-----------)..(....)...))))))))) ( -20.40) >DroSim_CAF1 131878 84 - 1 -UUGCACAAGGAAUAUGCCUAAUGAGCGCCAUUUUGCCGGCAUUUACUGCCA-AUGCCACACAAA-----------AAAGGACACAAGGGCGCUGUG -...(((.(((......)))....((((((.((.((((((((.....)))).-.((.....))..-----------...)).)).)).))))))))) ( -22.70) >DroEre_CAF1 136168 85 - 1 -UUGCACAAGGUAUAUGCCUAAUGAGCGCCAUUUUCAUGGCAUUUACUGCCAAAUGCCGCACAAAAAA-----------GGACACAAGGGCGCUGUC -...((..((((....))))..))((((((.......(((((.....)))))..((((..........-----------)).))....))))))... ( -23.20) >DroYak_CAF1 136584 95 - 1 -UUGCACAAGGAAUCUGCCUAAUGAGCGCCAUUUUCAUGGCAUUUACUGCCA-AUGCCGCACAAAAAAGGACAUAAAAAGGACACAAGGGCGCUGCA -.(((...(((......)))....((((((.......(((((.....)))))-(((((..........)).)))..............))))))))) ( -24.90) >DroAna_CAF1 131732 72 - 1 AGGACCAAAGGAAGCGCCUUAAUGAGCGCCAUUUUG-UGGCA-------------GCCACACAAAAGG-----------UAGAAGAGGGGCGCUUCA ..........((((((((((..(....(((.(((((-((...-------------....)))))))))-----------)...)..)))))))))). ( -27.40) >consensus _UUGCACAAGGAAUAUGCCUAAUGAGCGCCAUUUUGCCGGCAUUUACUGCCA_AUGCCACACAAAAAA________AAAGGACACAAGGGCGCUGCG ........(((......)))....((((((........(((((..........))))).....................(....)...))))))... (-16.43 = -16.08 + -0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:42 2006