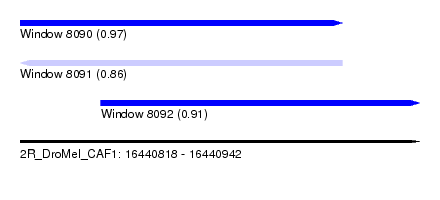
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,440,818 – 16,440,942 |
| Length | 124 |
| Max. P | 0.969913 |

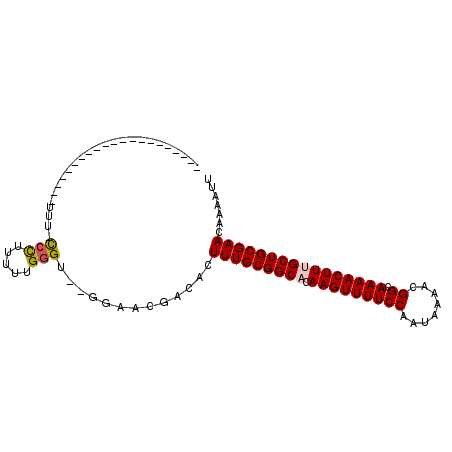
| Location | 16,440,818 – 16,440,918 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -19.91 |
| Energy contribution | -20.13 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16440818 100 + 20766785 CCCAUUUUUGUUUCUUUCUUUUUUUCCCUUUUUGGGU--GGAACGACACUUUGCGGCACAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU .....((((((((.......(((..(((.....))).--.))).........((((((.(((((((((........)).))))))))))))))))))))).. ( -27.00) >DroPse_CAF1 146269 80 + 1 ---------------------UUU-CCCAUUUUGGGGCAACAACGACACUUUGCGGCCGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU ---------------------..(-(((.....))))............((((((((.((((((((((........)).))))))))))))))))....... ( -23.10) >DroSec_CAF1 116310 99 + 1 CCCAUUUUUGUUUCUUUCUUUUUU-CCCUUUUUGGGU--GGAACGACACUUUGCGGCACAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU .....((((((((...((.(((..-(((.....))).--.))).))......((((((.(((((((((........)).))))))))))))))))))))).. ( -28.30) >DroEre_CAF1 116210 99 + 1 CCCAUUUUUGUUUCUUUCUUUUUU-CCUUUUUUGGGU--GGAACGACACUUUGCGGCACAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU .....((((((.((.((((....(-((......))).--)))).))....((((((((.(((((((((........)).))))))))))))))))))))).. ( -26.00) >DroMoj_CAF1 193841 76 + 1 ---------------------UUU-UGCUUUUUGGGC----GACGACACUUUGCGGCAGAAGUUUUCCAAUAAAAUGGCAAAACUUUGCUGCGAACAAAAUU ---------------------..(-((((.....)))----))......(((((((((((.((((((((......))).))))))))))))))))....... ( -23.50) >DroPer_CAF1 147304 80 + 1 ---------------------UUU-CCCAUUUUGGGGCAACAACGACACUUUGCGGCCGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU ---------------------..(-(((.....))))............((((((((.((((((((((........)).))))))))))))))))....... ( -23.10) >consensus _____________________UUU_CCCUUUUUGGGU__GGAACGACACUUUGCGGCACAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUU .........................(((.....))).............(((((((((.(((((((((........)).))))))))))))))))....... (-19.91 = -20.13 + 0.22)

| Location | 16,440,818 – 16,440,918 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.72 |
| Mean single sequence MFE | -21.73 |
| Consensus MFE | -14.85 |
| Energy contribution | -15.55 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16440818 100 - 20766785 AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUGUGCCGCAAAGUGUCGUUCC--ACCCAAAAAGGGAAAAAAAGAAAGAAACAAAAAUGGG ..((((((((((.((((((((((.(((......)))))))))).))).)).............--.(((.....)))...........).)))))))..... ( -23.00) >DroPse_CAF1 146269 80 - 1 AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUCGGCCGCAAAGUGUCGUUGUUGCCCCAAAAUGGG-AAA--------------------- ..........(((((((((((((.(((......))))))))).(((((.....).)))))))))))(((.....)))-...--------------------- ( -21.90) >DroSec_CAF1 116310 99 - 1 AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUGUGCCGCAAAGUGUCGUUCC--ACCCAAAAAGGG-AAAAAAGAAAGAAACAAAAAUGGG ..((((((((((.((((((((((.(((......)))))))))).))).)).............--.(((.....)))-..........).)))))))..... ( -23.00) >DroEre_CAF1 116210 99 - 1 AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUGUGCCGCAAAGUGUCGUUCC--ACCCAAAAAAGG-AAAAAAGAAAGAAACAAAAAUGGG ..((((((((((.((((((((((.(((......)))))))))).))).))......((.((((--..........))-))....))..).)))))))..... ( -21.60) >DroMoj_CAF1 193841 76 - 1 AAUUUUGUUCGCAGCAAAGUUUUGCCAUUUUAUUGGAAAACUUCUGCCGCAAAGUGUCGUC----GCCCAAAAAGCA-AAA--------------------- ..(((((((.((.((((((((((.(((......)))))))))).))).))...(((....)----))......))))-)))--------------------- ( -19.00) >DroPer_CAF1 147304 80 - 1 AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUCGGCCGCAAAGUGUCGUUGUUGCCCCAAAAUGGG-AAA--------------------- ..........(((((((((((((.(((......))))))))).(((((.....).)))))))))))(((.....)))-...--------------------- ( -21.90) >consensus AAUUUUGUUCGCAGCAAAGUUUUGCCGUUUUAUUGGAAAACUUCUGCCGCAAAGUGUCGUUCC__ACCCAAAAAGGG_AAA_____________________ .(((((((..(((...(((((((.(((......)))))))))).))).)))))))...........(((.....)))......................... (-14.85 = -15.55 + 0.70)

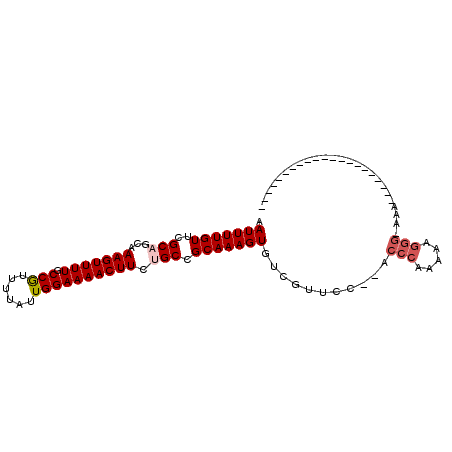
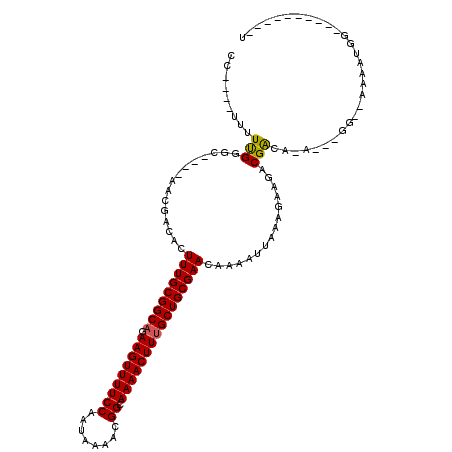
| Location | 16,440,843 – 16,440,942 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.19 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16440843 99 + 20766785 CCC---UUUUUGGGU--GGAACGACACUUUGCGGCACAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGACA-A---GG--AAAAUGG----------U .((---((.((((((--(......))))((((((((.(((((((((........)).)))))))))))))))................))).)-)---))--.......----------. ( -27.70) >DroVir_CAF1 167648 95 + 1 --------UUUGGGC----GACGACACUUUGCGGCAGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGAC-GACAACG--ACAACGA----------C --------(((((..----..).....(((((((((((.(((((((........)).))))))))))))))))))))...............-.......--.......----------. ( -20.40) >DroGri_CAF1 136410 82 + 1 --------UUUGGGC----GACAACACUUUGCGGCAGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGACA-A------------------------- --------(((((..----..).....(((((((((((.(((((((........)).))))))))))))))))))))................-.------------------------- ( -20.40) >DroWil_CAF1 265930 110 + 1 CCUCUCUUUUUGGG----CAACGACACUUUGCGGCAGAAGUUUUCCAAUAAAAUGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGGCA-A---GG--AAAAUGUAUCCACAAGUG .((.(((((((.(.----...).....(((((((((((.((((((((......))).))))))))))))))))........))))))).))..-.---((--(.......)))....... ( -28.30) >DroAna_CAF1 111560 103 + 1 CCU-UUUUAUCGGGC--GGGACGACACUUUGCGGCAGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGACA-A---GGCAAAAAUGG----------U (((-((((..(..((--((.....)..(((((((((((.(((((((........)).))))))))))))))))...............)).).-.---)..))))).))----------. ( -23.80) >DroPer_CAF1 147307 101 + 1 CCC---AUUUUGGGGCAACAACGACACUUUGCGGCCGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGACA-A---GG--AAAAUGC----------U (((---.....)))(((..........((((((((.((((((((((........)).))))))))))))))))...............(....-.---).--....)))----------. ( -23.80) >consensus CC____UUUUUGGGC____AACGACACUUUGCGGCAGAAGUUUUCCAAUAAAACGGCAAAACUUUGCUGCGAACAAAAUUAAAGAAGACGACA_A___GG__AAAAUGG__________U .........(((...............(((((((((.(((((((((........)).))))))))))))))))...............)))............................. (-18.30 = -18.19 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:32 2006