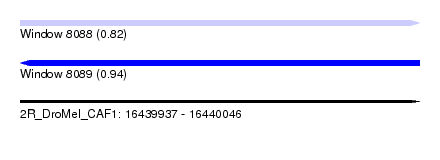
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,439,937 – 16,440,046 |
| Length | 109 |
| Max. P | 0.942596 |

| Location | 16,439,937 – 16,440,046 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -26.17 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.52 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
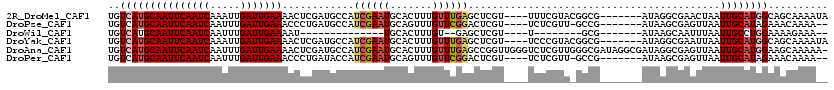
>2R_DroMel_CAF1 16439937 109 + 20766785 UAUUUUGCUGCCAUGCAAUUAGUUCGCCUAU-------CGCCGUACGAAA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACA ...........(((((((.......(((.((-------((..((((....----...............))))...)))))))........(((((((.....))))))))))))))... ( -26.51) >DroPse_CAF1 145192 106 + 1 --UUUUGUUUCUAUGCAAUUAACUCGCUUAU-------CGGC-AACGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGCAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACA --.........(((((((((.(((((....(-------((..-..)))..----.)))))..((((....(((........))).....))))(((((.....))))))))))))))... ( -26.40) >DroWil_CAF1 265217 83 + 1 --UUUCUUUUCCAGGCAAUUAAAUUGCUUAU-------CGC--------A----ACGAGCUC--ACAAAGUGCA--------------AUUUUCAAUCAAAUUGAUUGAAUUGCAUGACA --..........(((((((...)))))))..-------.((--------.----....))..--.....(((((--------------((..((((((.....))))))))))))).... ( -18.10) >DroYak_CAF1 116330 109 + 1 UAUUUUGCUGCCAUGCAAUUAAUUCGCCUAU-------CGCCGUACGGGA----ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAUUUGAUUGAAUUGCAUGACA ...........(((((((..((((((.((((-------((..(((((...----.)(....).......))))...))))))...))))))(((((((.....))))))))))))))... ( -28.50) >DroAna_CAF1 110568 119 + 1 -UUUUUGCUUCCAUGCAAUUAACUCGCCUAUCGCCUAUCGCCCAACGAGACCCAACCGGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAAUUGAUUGAAUUGCAUGACA -..........(((((((..((((((.((((((.....(((.....(((.((.....))))).......)))....))))))...))))))(((((((.....))))))))))))))... ( -31.30) >DroPer_CAF1 146240 106 + 1 --UUUUGUUUCUAUGCAAUUAACUCGCUUAU-------CGGC-AACGAGA----ACGAGUCCGAACAAACUGCAUUCGAUGGUAUCAGGGUUUCAAUCAAAUUGAUUGAAUUGCAUGACA --.........(((((((((.(((((....(-------((..-..)))..----.)))))..((((...(((.((........))))).))))(((((.....))))))))))))))... ( -26.20) >consensus __UUUUGCUUCCAUGCAAUUAACUCGCCUAU_______CGCC_AACGAGA____ACGAGCUCAAACAAAGUGCAUUCGAUGGCAUCGAGUUUUCAAUCAAAUUGAUUGAAUUGCAUGACA ...........(((((((.......(((..........((.....))...........((...........)).......)))........(((((((.....))))))))))))))... (-14.24 = -14.52 + 0.28)

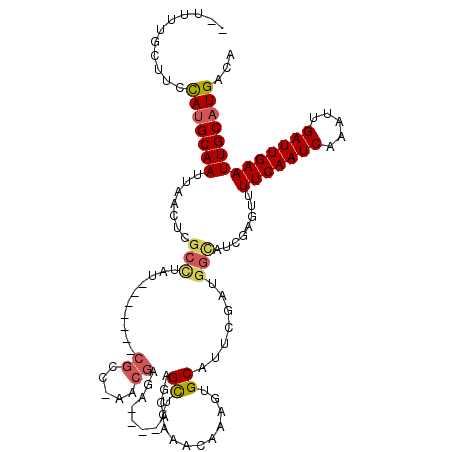
| Location | 16,439,937 – 16,440,046 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -15.30 |
| Energy contribution | -16.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16439937 109 - 20766785 UGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UUUCGUACGGCG-------AUAGGCGAACUAAUUGCAUGGCAGCAAAAUA (((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......))))).-------...))))......))))))))))........ ( -34.10) >DroPse_CAF1 145192 106 - 1 UGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUGCCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCGUU-GCCG-------AUAAGCGAGUUAAUUGCAUAGAAACAAAA-- ..(((((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((..-..))-------)..))))))))..)))))).)).......-- ( -26.40) >DroWil_CAF1 265217 83 - 1 UGUCAUGCAAUUCAAUCAAUUUGAUUGAAAAU--------------UGCACUUUGU--GAGCUCGU----U--------GCG-------AUAAGCAAUUUAAUUGCCUGGAAAAGAAA-- ..(((.(((((((((((.....))))))((((--------------(((...(((.--.(.....)----.--------.))-------)...))))))).))))).)))........-- ( -20.40) >DroYak_CAF1 116330 109 - 1 UGUCAUGCAAUUCAAUCAAAUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU----UCCCGUACGGCG-------AUAGGCGAAUUAAUUGCAUGGCAGCAAAAUA (((((((((((((((((.....))))))).......((((.((((((.......))))))((.(((----......))))).-------...))))......))))))))))........ ( -34.10) >DroAna_CAF1 110568 119 - 1 UGUCAUGCAAUUCAAUCAAUUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCCGGUUGGGUCUCGUUGGGCGAUAGGCGAUAGGCGAGUUAAUUGCAUGGAAGCAAAAA- ..(((((((((((((((.....)))))))(((((...(((((((..(((.(((....((((((.....)).))))..))).).))..)))).))))))))..)))))))).........- ( -36.90) >DroPer_CAF1 146240 106 - 1 UGUCAUGCAAUUCAAUCAAUUUGAUUGAAACCCUGAUACCAUCGAAUGCAGUUUGUUCGGACUCGU----UCUCGUU-GCCG-------AUAAGCGAGUUAAUUGCAUAGAAACAAAA-- ..(((((((((((((((.....))))))).............(((((.......)))))(((((((----(.(((..-..))-------)..))))))))..)))))).)).......-- ( -26.40) >consensus UGUCAUGCAAUUCAAUCAAUUUGAUUGAAAACUCGAUGCCAUCGAAUGCACUUUGUUUGAGCUCGU____UCUCGUU_GGCG_______AUAAGCGAGUUAAUUGCAUGGAAACAAAA__ ..(((((((((((((((.....)))))))............((((((.......))))))..........................................)))))))).......... (-15.30 = -16.05 + 0.75)
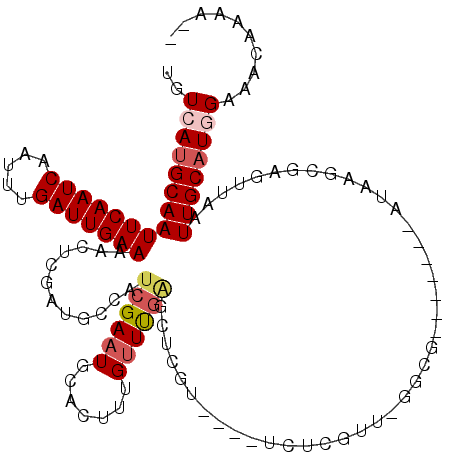
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:29 2006