
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,436,183 – 16,436,277 |
| Length | 94 |
| Max. P | 0.555207 |

| Location | 16,436,183 – 16,436,277 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -18.82 |
| Consensus MFE | -13.89 |
| Energy contribution | -14.03 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.82 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
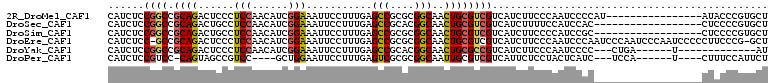
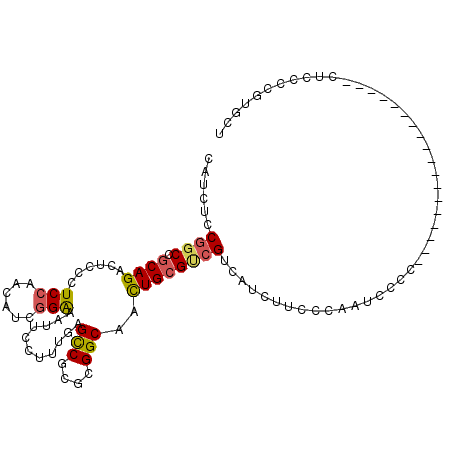
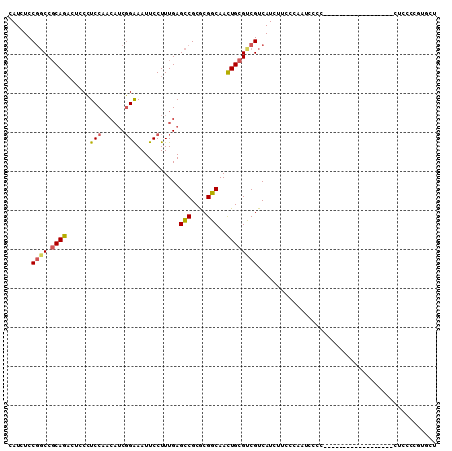
>2R_DroMel_CAF1 16436183 94 - 20766785 CAUCUCCGGCCGCAGACUCCCUCCAACAUCGGAAAUUCCUUUGAGCCGCGCGGCAACUGCGUCGUCAUCUUCCCAAUCCCCAU----------------AUACCCGUGCU ......((((.((((...........((..((.....))..)).(((....)))..))))))))...................----------------........... ( -18.00) >DroSec_CAF1 111752 92 - 1 CAUCUCCGGCCGCAGACUGCCUCCAACAUCGGAAAUUCCUUUGAGCCGCACGGCAACUGCGUCGUCAUCUUUUCCAUCCAC------------------CUCCCCGUGCU ......((((.((((...........((..((.....))..)).(((....)))..))))))))..............(((------------------......))).. ( -18.50) >DroSim_CAF1 107474 92 - 1 CAUCUCCGGCCGCAGACUGCCUCCAACAUCGGAAAUUCCUUUGAGCCGCGCGGCAACUGCGUCGUCAUCUUCCCCAUCCGC------------------CUCCCCGUGCU .......(((..((((.....(((......)))......)))).)))((((((.....(((.................)))------------------....)))))). ( -19.73) >DroEre_CAF1 111632 108 - 1 CAUCUCC-GCCGCAGACUCCCUCCAACAUCGGAAAUUCCUUUGAGCCGCGCGGCAACUGCGUCGUCAUCUUCCCAAUCCCAAUCCCAAUCCCAAUCCCCCUUCCCG-GCU .......-(((((((...........((..((.....))..)).(((....)))..))))(....).......................................)-)). ( -17.60) >DroYak_CAF1 112626 88 - 1 CAUCUCCGGCCGCAGACUCCCUCCAACAUCGGAAAUUCCUUUGAGCCGCACGGCAACUGCGCCGUCAUCUUCCCAAUCCCC---CUGA------U-------------AU ......((((.((((...........((..((.....))..)).(((....)))..)))))))).................---....------.-------------.. ( -20.70) >DroPer_CAF1 142168 92 - 1 CAUCUCCGUCC-CAGUAGCCGUCC----GCUGGAAUUCCUUUGAGUCGCGCGGCAAUUGCGUCGUCAUUCUCCUACUCAUC---UCCA------U----CUUUCCAUUCU ......((.(.-((((.(((((.(----(((.(((....))).)).)).))))).)))).).)).................---....------.----........... ( -18.40) >consensus CAUCUCCGGCCGCAGACUCCCUCCAACAUCGGAAAUUCCUUUGAGCCGCGCGGCAACUGCGUCGUCAUCUUCCCAAUCCCC__________________CUCCCCGUGCU ......((((.((((......(((......)))...........(((....)))..)))))))).............................................. (-13.89 = -14.03 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:27 2006