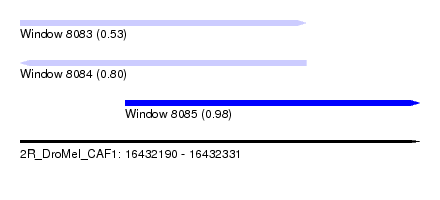
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,432,190 – 16,432,331 |
| Length | 141 |
| Max. P | 0.975859 |

| Location | 16,432,190 – 16,432,291 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -20.62 |
| Energy contribution | -22.66 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531089 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
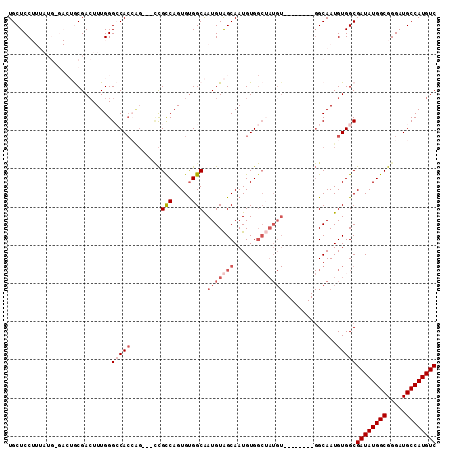
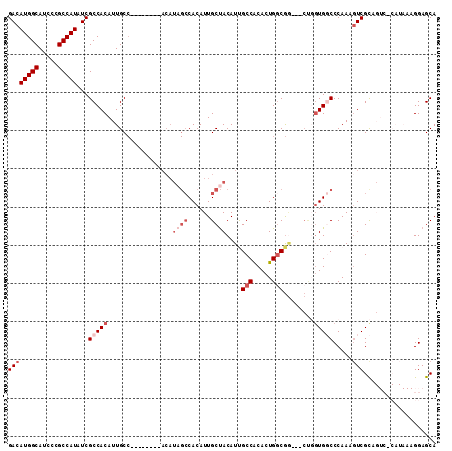
>2R_DroMel_CAF1 16432190 101 + 20766785 UGCUCCUUUAUGCGACUGCGACUUUGGGCCAACAG---CCGUCAGUGUGGCAAUGUAGCAAUGUGGCUAUGU--------GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC .((..(....(((.(((((.((.((((((.....)---))..))).)).)))..(((((......)))))))--------.)))..)..))((((((((.....)))))))) ( -34.10) >DroSec_CAF1 107811 101 + 1 UGCUCCUUUAUGGGACUGCGACUUUGGGCCACCAG---CCGCCAGUGUGGCAAUGUAGCAAUGUGGCUAUGU--------GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC .((((((....))))..))........(((((..(---(((((.....))).(((((((......)))))))--------)))...)))))((((((((.....)))))))) ( -40.20) >DroSim_CAF1 103559 101 + 1 UGCUCCUUUAUGCGACUGCGACUUUGGGCCACCAG---CCGCCAGUGUGGCAAUGUAGCAAUGUGGCUAUGU--------GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC .((........))....((.((.....(((((..(---((((....)))))...(((((......)))))))--------)))...)).))((((((((.....)))))))) ( -38.30) >DroEre_CAF1 107684 78 + 1 UGCUCC-----------ACGACUUUGGGGCAC-------UGCCAGUGCGGCGAUGUGGCAAUGUG----------------GCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC ((((((-----------(......))))))).-------.((((.(((.(((.((...)).))).----------------)))...))))((((((((.....)))))))) ( -32.40) >DroYak_CAF1 108452 101 + 1 UGCUCC-----------ACGAGUUUGGGGCACGGGGCAUCGCCAGUGUGGCGAUGUAGCAAUGUGGCAAUGUGGCAUUGUGGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC .(((((-----------(......))))))(((.((((((((((.(((.(((.(((.(((((((.((...)).))))))).))).))).)))...))))..)))))).))). ( -45.50) >consensus UGCUCCUUUAUG_GACUGCGACUUUGGGCCACCAG___CCGCCAGUGUGGCAAUGUAGCAAUGUGGCUAUGU________GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUC ...........................(((((........(((.....))).(((((((......)))))))..............)))))((((((((.....)))))))) (-20.62 = -22.66 + 2.04)

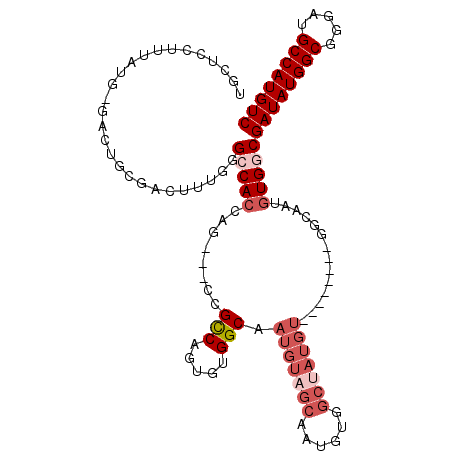
| Location | 16,432,190 – 16,432,291 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -16.06 |
| Energy contribution | -18.06 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
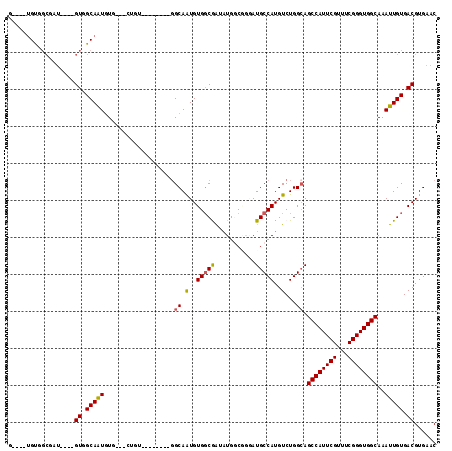
>2R_DroMel_CAF1 16432190 101 - 20766785 GACAUGGCAUCCCGCCAUAUCGCCACAUUGCC--------ACAUAGCCACAUUGCUACAUUGCCACACUGACGG---CUGUUGGCCCAAAGUCGCAGUCGCAUAAAGGAGCA ((((((((.....)))))...((.((...(((--------(.((((((.....((......))(.....)..))---)))))))).....)).)).)))((........)). ( -27.30) >DroSec_CAF1 107811 101 - 1 GACAUGGCAUCCCGCCAUAUCGCCACAUUGCC--------ACAUAGCCACAUUGCUACAUUGCCACACUGGCGG---CUGGUGGCCCAAAGUCGCAGUCCCAUAAAGGAGCA ((((((((.....)))))...(((((...(((--------...((((......))))....(((.....)))))---)..))))).....)))((..(((......))))). ( -36.10) >DroSim_CAF1 103559 101 - 1 GACAUGGCAUCCCGCCAUAUCGCCACAUUGCC--------ACAUAGCCACAUUGCUACAUUGCCACACUGGCGG---CUGGUGGCCCAAAGUCGCAGUCGCAUAAAGGAGCA ((((((((.....)))))...(((((...(((--------...((((......))))....(((.....)))))---)..))))).....)))((....))........... ( -34.00) >DroEre_CAF1 107684 78 - 1 GACAUGGCAUCCCGCCAUAUCGCCACAUUGC----------------CACAUUGCCACAUCGCCGCACUGGCA-------GUGCCCCAAAGUCGU-----------GGAGCA ((.(((((.....))))).))((..((((((----------------((...(((.........))).)))))-------)))..(((......)-----------)).)). ( -23.50) >DroYak_CAF1 108452 101 - 1 GACAUGGCAUCCCGCCAUAUCGCCACAUUGCCACAAUGCCACAUUGCCACAUUGCUACAUCGCCACACUGGCGAUGCCCCGUGCCCCAAACUCGU-----------GGAGCA ((.(((((.....))))).))((..(((((...)))))((((...((......))..(((((((.....))))))).................))-----------)).)). ( -29.50) >consensus GACAUGGCAUCCCGCCAUAUCGCCACAUUGCC________ACAUAGCCACAUUGCUACAUUGCCACACUGGCGG___CUGGUGGCCCAAAGUCGCAGUC_CAUAAAGGAGCA ((((((((.....)))))...(((((.................((((......))))....(((.....)))........))))).....)))................... (-16.06 = -18.06 + 2.00)

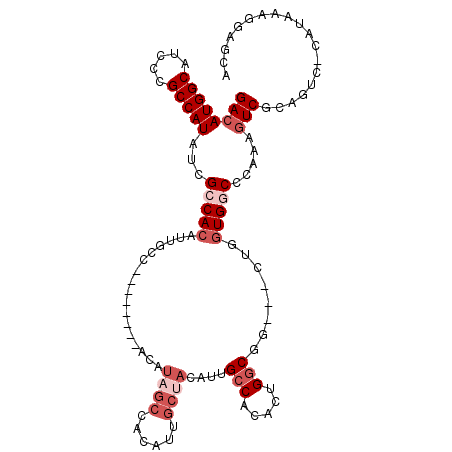
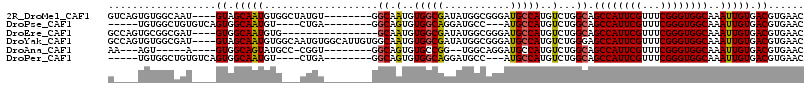
| Location | 16,432,227 – 16,432,331 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -24.34 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
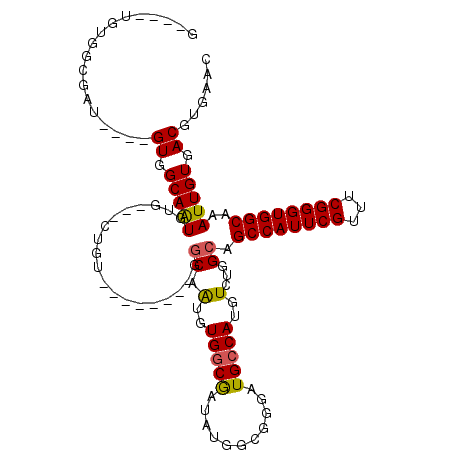
>2R_DroMel_CAF1 16432227 104 + 20766785 GUCAGUGUGGCAAU----GUAGCAAUGUGGCUAUGU--------GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC ((((((((((((..----...((.((((.(((((((--------....))))))).)))).))....))))))).))))).((((((((...))))))))................ ( -40.50) >DroPse_CAF1 137238 96 + 1 -----UGUGGCUGUGUCAGUGGCAAUGU----CUGA--------GGCAGUGUGGCAGGAUGCC---AUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC -----(((.((((...)))).)))((((----(...--------(.((((((((((((...))---.))))))).))).).((((((((...)))))))).......))))).... ( -36.40) >DroEre_CAF1 107706 96 + 1 GCCAGUGCGGCGAU----GUGGCAAUGUG----------------GCAAUGUGGCGAUAUGGCGGGAUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC ((((..((.(((.(----((.((...)).----------------))).))).))((((((((.....)))))))))))).((((((((...))))))))................ ( -38.90) >DroYak_CAF1 108481 112 + 1 GCCAGUGUGGCGAU----GUAGCAAUGUGGCAAUGUGGCAUUGUGGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUCUGGGAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC (((.....)))..(----((.(((((((.((...)).))))))).)))((((.((((((((((.....)))))))).....((((((((...)))))))).....)).)))).... ( -44.50) >DroAna_CAF1 103055 93 + 1 AA---AGU-----A----GUGGCAGUAUGCC-CGGU--------GGCAGUGUGCCGG--UGGCAGGAUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC ..---...-----.----((.(((((.((((-.(((--------((((.(.((((..--.)))).).))))))..).))))((((((((...))))))))..))))).))...... ( -37.60) >DroPer_CAF1 138334 96 + 1 -----UGUGGCUGUGUCAGUGGCAAUGU----CUGA--------GGCAGUGUGGCAGGAUGCC---AUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC -----(((.((((...)))).)))((((----(...--------(.((((((((((((...))---.))))))).))).).((((((((...)))))))).......))))).... ( -36.40) >consensus G____UGUGGCGAU____GUGGCAAUGUG___CUGU________GGCAAUGUGGCGAUAUGGCGGGAUGCCAUGUCUGGCAGCCAUUCGUUUCGGGUGGCAAAUUGUGACGUGAAC ..................((.(((((...................((.(..(((((...........)))))..)...)).((((((((...))))))))..))))).))...... (-24.34 = -24.07 + -0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:26 2006