| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,431,619 – 16,431,714 |
| Length | 95 |
| Max. P | 0.902302 |

| Location | 16,431,619 – 16,431,714 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -12.77 |
| Energy contribution | -13.43 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
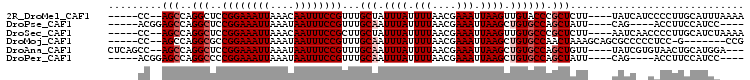
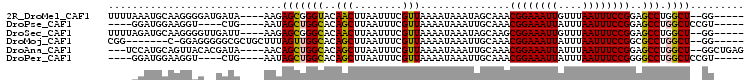
>2R_DroMel_CAF1 16431619 95 + 20766785 -----CC--AGCCAGGCUCCGGAAAUUAAACAAUUUCCGUUUGCUAUUUAUUUUAACGAAAUUAAGUUGUACCCGCUCUU----UAUCAUCCCCUUGCAUUUAAAA -----..--.((.(((...((((((((....))))))))..(((.(((((.(((....))).))))).))).........----........))).))........ ( -15.60) >DroPse_CAF1 136583 89 + 1 -----ACGGAGCCAGGCUCCGGAAAUUAAAUAAUUUCCGUUUGCAAUUUAUUUUAACGAAAUUAAGCUGUGCCAGCUAUU----CAG----ACCUUCCAUCC---- -----..(((((...)))))((((......(((((((.(((.............))))))))))(((((...)))))...----...----...))))....---- ( -20.22) >DroSec_CAF1 107229 95 + 1 -----CC--AGCCAGGCUCCGGAAAUUAAACAAUUUCCGCUUGCUAUUUAUUUUAACGAAAUUAAGUUGUGCCCGCUCUU----AAUCAACCCCUUGCAUCUAAAA -----..--.((.(((....(((((((....)))))))((..((.(((((.(((....))).)))))...))..))....----........))).))........ ( -16.60) >DroMoj_CAF1 179975 91 + 1 -----CC--AGCCAGGCGCCGGAAAUUAAAUAAUUUCCGUUUGCAAUUUAUUUUAACGAAAUUAAGCUGUGCCAACUAAAGCAGCGCCCCCUCC-G-------CCG -----..--.....(((((((((((((....))))))))...)).....................(((((..........))))).........-)-------)). ( -21.30) >DroAna_CAF1 102552 97 + 1 CUCAGCC--AGCCAGGCUCCGGAAAUUAAAUAAUUUCCGUUUGCAAUUUAUUUUAACGAAAUUAAGCUGUGCCAGCUGUU----UAUCGUGUAACUGCAUGGA--- .....((--(..(((((..((((((((....))))))))...))...........((((.....(((((...)))))...----..))))....)))..))).--- ( -25.50) >DroPer_CAF1 137679 89 + 1 -----ACGGAGCCAGGCCCCGGAAAUUAAAUAAUUUCCGUUUGCAAUUUAUUUUAACGAAAUUAAGCUGUGCCAGCUAUU----CAG----ACCUUCCAUCC---- -----..((((....((..((((((((....))))))))...)).............(((....(((((...))))).))----)..----...))))....---- ( -18.00) >consensus _____CC__AGCCAGGCUCCGGAAAUUAAAUAAUUUCCGUUUGCAAUUUAUUUUAACGAAAUUAAGCUGUGCCAGCUAUU____CAUC__CACCUUGCAUCC____ .........(((..(((..((((((((....))))))))...(((.((((.(((....))).)))).)))))).)))............................. (-12.77 = -13.43 + 0.67)
| Location | 16,431,619 – 16,431,714 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -15.05 |
| Energy contribution | -14.88 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
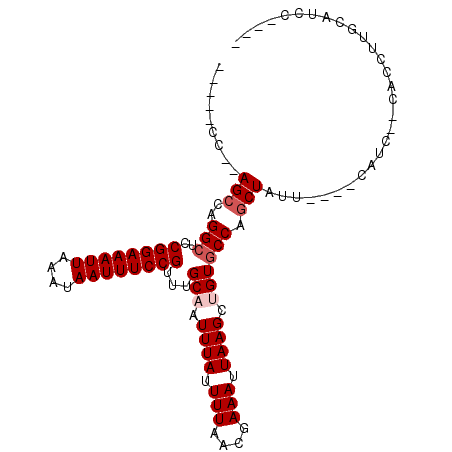
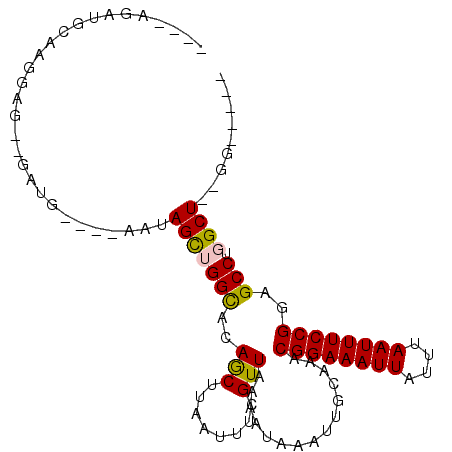
>2R_DroMel_CAF1 16431619 95 - 20766785 UUUUAAAUGCAAGGGGAUGAUA----AAGAGCGGGUACAACUUAAUUUCGUUAAAAUAAAUAGCAAACGGAAAUUGUUUAAUUUCCGGAGCCUGGCU--GG----- ........((.(((........----...((((((...........))))))...............((((((((....))))))))...))).)).--..----- ( -16.70) >DroPse_CAF1 136583 89 - 1 ----GGAUGGAAGGU----CUG----AAUAGCUGGCACAGCUUAAUUUCGUUAAAAUAAAUUGCAAACGGAAAUUAUUUAAUUUCCGGAGCCUGGCUCCGU----- ----(((.(..((((----..(----((.(((((...)))))....)))..................((((((((....))))))))..))))..))))..----- ( -22.50) >DroSec_CAF1 107229 95 - 1 UUUUAGAUGCAAGGGGUUGAUU----AAGAGCGGGCACAACUUAAUUUCGUUAAAAUAAAUAGCAAGCGGAAAUUGUUUAAUUUCCGGAGCCUGGCU--GG----- ......................----...(((((((.............((((.......))))...((((((((....))))))))..)))).)))--..----- ( -18.60) >DroMoj_CAF1 179975 91 - 1 CGG-------C-GGAGGGGGCGCUGCUUUAGUUGGCACAGCUUAAUUUCGUUAAAAUAAAUUGCAAACGGAAAUUAUUUAAUUUCCGGCGCCUGGCU--GG----- (((-------(-...(((.((((((((......)))..)))..........................((((((((....)))))))))).))).)))--).----- ( -26.70) >DroAna_CAF1 102552 97 - 1 ---UCCAUGCAGUUACACGAUA----AACAGCUGGCACAGCUUAAUUUCGUUAAAAUAAAUUGCAAACGGAAAUUAUUUAAUUUCCGGAGCCUGGCU--GGCUGAG ---....(((((((..((((..----...(((((...))))).....)))).......)))))))..((((((((....)))))))).((((.....--))))... ( -27.00) >DroPer_CAF1 137679 89 - 1 ----GGAUGGAAGGU----CUG----AAUAGCUGGCACAGCUUAAUUUCGUUAAAAUAAAUUGCAAACGGAAAUUAUUUAAUUUCCGGGGCCUGGCUCCGU----- ----(((.(..((((----(((----((.(((((...)))))....)))..................((((((((....))))))))))))))..))))..----- ( -26.80) >consensus ____AGAUGCAAGGAG__GAUG____AAUAGCUGGCACAGCUUAAUUUCGUUAAAAUAAAUUGCAAACGGAAAUUAUUUAAUUUCCGGAGCCUGGCU__GG_____ .............................(((((((..(((........)))...............((((((((....))))))))..))).))))......... (-15.05 = -14.88 + -0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:23 2006