| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,423,101 – 16,423,316 |
| Length | 215 |
| Max. P | 0.869180 |

| Location | 16,423,101 – 16,423,210 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -30.04 |
| Consensus MFE | -24.46 |
| Energy contribution | -24.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.646684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16423101 109 - 20766785 UAUUUUCUAAAAACAUUGCAUUGGCCAUUGGAGGUGGAUGCCCUGGAAGGCCACAUCCCACUCGGACACAUCCCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC ..............(((((.(((((....((((((((..(((......)))....(((.....)))......))))))))((((......)))).)))))))))).... ( -31.70) >DroSec_CAF1 98967 109 - 1 AAUUUUCUAAAAACAUUGCAUUGGCCAUUGGAGGUGGAUGCCCUGGAAGGCCGCAUCCCACUCCGCCACAUCCCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC ..............(((((.(((((....(((((((((((...((((.((.......))..))))...))).))))))))((((......)))).)))))))))).... ( -31.80) >DroSim_CAF1 94737 109 - 1 AAUUUUCUAAAAACAUUGCAUUGGCCAUUGGAGGUGGAUGCCCUGGGAGGCCACAUCCCACUCCGCCACAUCCCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC ..............(((((.(((((....(((((((((((.....((((...........))))....))).))))))))((((......)))).)))))))))).... ( -33.00) >DroEre_CAF1 98571 93 - 1 A-UUUUCCAAAAAUAUUGCAUUGGUCAUUGAAGGUGGAUGCCCUGGAAGGCCACAUC---------------CCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC .-...........((((((.(((((....((((..(((((...(((....)))))))---------------)..)))).((((......)))).)))))))))))... ( -24.30) >DroYak_CAF1 99076 101 - 1 AAAUUUCUAAAAAUAUUGCAUUGGUCAUUGGAGGUGGAUGCCCUGGAAGGCCAUAUCCCGCU--------UCCCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC .............((((((.(((((....((((((((..((...(((........))).)).--------..))))))))((((......)))).)))))))))))... ( -29.40) >consensus AAUUUUCUAAAAACAUUGCAUUGGCCAUUGGAGGUGGAUGCCCUGGAAGGCCACAUCCCACUC_G_CACAUCCCACUUCCGACACUUGAUUGUCAGCCGGGUAAUAGAC ..............(((((.(((((....((((((((..(((......))).....................))))))))((((......)))).)))))))))).... (-24.46 = -24.42 + -0.04)

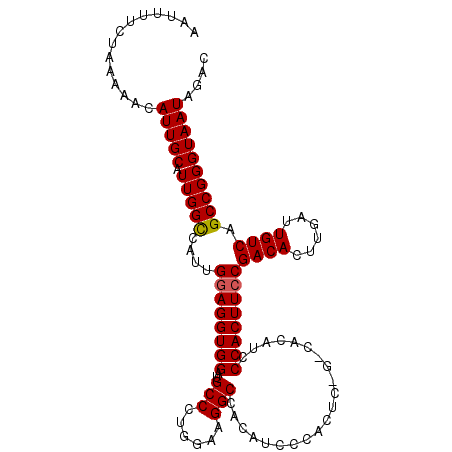
| Location | 16,423,210 – 16,423,316 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.69 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -5.98 |
| Energy contribution | -5.30 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.869180 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16423210 106 + 20766785 AUUCAGCCAACGAUACAAACUUGUCGCGUAAUUUAAACAUUUUGGGCU--------------UUUAAGUAGCUACUCCAUAUACAUACAUGAUUUGCUAGAAAUGUACAGCUGAAUUUGG (((((((...(((((......)))))..........(((((((.(((.--------------.(((.(((...............))).)))...))).)))))))...))))))).... ( -20.96) >DroSec_CAF1 99076 102 + 1 AUUCAGCCAACGAUACAAACUUGUCUCGUAAUUAUAACAUUUAAGGCU--------------AAUAAGUAGCCACUGCAGAU----ACAUGAUUUGCUAAAAAUGUACAGCUGAAUUUGG (((((((..((((.(((....))).)))).......((((((..((((--------------(.....)))))...((((((----.....))))))...))))))...))))))).... ( -27.80) >DroSim_CAF1 94846 102 + 1 AUUCAGCCAACGAUACAAACUUGUCUCGUAAUUAAAACAUUUAAGGCU--------------AAUAAGUAGCUACUGCAGAU----ACAUGAUUUGCUACAAAUGUACAGCUGAAUUUGG (((((((..((((.(((....))).)))).......((((((..((((--------------(.....)))))...((((((----.....))))))...))))))...))))))).... ( -25.20) >DroEre_CAF1 98664 115 + 1 AUUCAGUCAACGACACAAACUUGUCUCGCAACUAAAGCAUUUAAGGCUGACUUCCCACAGCAAAUAAGUAGCUUUU-UACUU----ACAUGAUUUGUUGGAAAUUUCCAGUUUAAUUUGA ..((((((...((((......))))..((.......))......)))))).((((.(((.((..(((((((....)-)))))----)..))...))).)))).................. ( -22.60) >DroYak_CAF1 99177 110 + 1 AUUAAGCCAACCAUACUAACUUGUCUUGUAAUUAAAACAUUUUAGGCUGAUUUUACUCAACGAAUAAGUAGCUACUACAGAU----ACAUGAUUUGAUAGAAAUUUGCAUGGUA------ .........(((((.((((..(((.(((....))).)))..))))((.(((((((((.........))))....((((((((----.....))))).)))))))).))))))).------ ( -16.80) >consensus AUUCAGCCAACGAUACAAACUUGUCUCGUAAUUAAAACAUUUAAGGCU______________AAUAAGUAGCUACUGCAGAU____ACAUGAUUUGCUAGAAAUGUACAGCUGAAUUUGG ....((((...((((......))))..((.......))......))))....................((((.....((..........))....))))..................... ( -5.98 = -5.30 + -0.68)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:17 2006