
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,414,234 – 16,414,382 |
| Length | 148 |
| Max. P | 0.998243 |

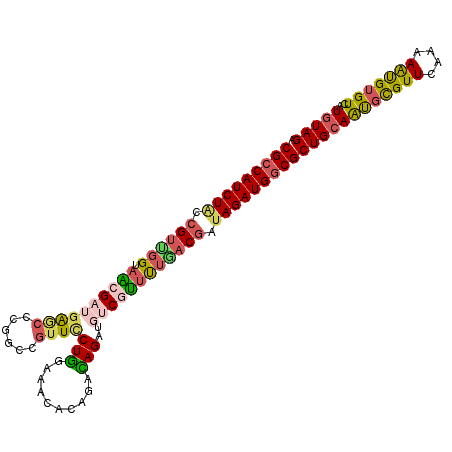
| Location | 16,414,234 – 16,414,342 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.82 |
| Mean single sequence MFE | -36.28 |
| Consensus MFE | -22.50 |
| Energy contribution | -23.30 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16414234 108 - 20766785 CGCCAUCUACCGUUGGUAACGAUGAGCCCGGCCGUUCCUGGAAACACAGGCAGAUCUCGUUUUGACGAUAGAUGUCGCUGCAAUGUGUUCAAAAGCGUGUCAUUUAGA (((((((((.(((..(.(((((.((.....(((.....((....))..)))...))))))))..))).)))))).(((......))).......)))........... ( -29.90) >DroSec_CAF1 90155 108 - 1 CGCCAUCUACAGUUGGUAACGAUGAGCCCGGACGUUCCUGGAAACACAGGUAGAUGUCGUUUUAACGAUAGAUGGCGCUGCAAUUCGUUCAAAAAUGUGUCAUGUAGA (((((((((..(((((.(((((((.(((.((.....))((....))..)))...))))))))))))..)))))))))(((((...((((....)))).....))))). ( -34.20) >DroSim_CAF1 85878 108 - 1 CGCCAUCUACCGUUGGUAACGAUGAGCGCGGUCGUUUCUGGAAACACAAACAGAUGUCGUUUUGACGACAGAUGGCGCUGCAAUGCGUUCAAAAAUGUGUCAUGUAGA ((((((((..(((..(.((((((.......(((((((.((....)).)))).))))))))))..)))..))))))))((((((((((((....)))))))..))))). ( -37.81) >DroEre_CAF1 89610 107 - 1 CGCCAUCUGUCGUCCGUAGAG-UGGUCCCCGCGGUUUCUACAAACACGGAUAGGUGUCGCCUUGCCGAUAGAUGGCGCUGCAGUGCGUUCAAAAGUGUGUCAUGUAGA (((((((((((((((((...(-(((...)))).((((....)))))))))).((((......)))))))))))))))(((((..(((..(....)..)))..))))). ( -40.30) >DroYak_CAF1 89872 105 - 1 CGCCAUCUACCGUCGGUAGAG---GAAGCCGUGGUUACUGGAAACACAUACAGGUGUCGUUCUCACGAUAGAUGGCGCUGCAGCGCGUUUAAAAAUGUGUCAUGUAGA ((((((((...((((..((((---((.(((...((...((....))...)).))).)).))))..))))))))))))((((((((((((....)))))))..))))). ( -39.20) >consensus CGCCAUCUACCGUUGGUAACGAUGAGCCCGGCCGUUCCUGGAAACACAGACAGAUGUCGUUUUGACGAUAGAUGGCGCUGCAAUGCGUUCAAAAAUGUGUCAUGUAGA (((((((((.((((((.((((((((((......))))(((..........)))..)))))))))))).)))))))))((((((((((((....)))))))..))))). (-22.50 = -23.30 + 0.80)

| Location | 16,414,262 – 16,414,382 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -24.99 |
| Energy contribution | -27.42 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.04 |
| SVM RNA-class probability | 0.998243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16414262 120 + 20766785 CAGCGACAUCUAUCGUCAAAACGAGAUCUGCCUGUGUUUCCAGGAACGGCCGGGCUCAUCGUUACCAACGGUAGAUGGCGCCAGUGCAACAUUAAGGACCUUUUCCAAUUUCAAAUCAUA ..(((.((((((((((...(((((.....((((((((((....)))))..)))))...)))))....)))))))))).)))..............(((.....))).............. ( -35.50) >DroSec_CAF1 90183 99 + 1 CAGCGCCAUCUAUCGUUAAAACGACAUCUACCUGUGUUUCCAGGAACGUCCGGGCUCAUCGUUACCAACUGUAGAUGGCGCCAGUGCGACAUCA-------UCUCA-------------- ..(((((((((((.(((..(((((......(((((((((....)))))..))))....)))))...))).))))))))))).............-------.....-------------- ( -31.30) >DroSim_CAF1 85906 106 + 1 CAGCGCCAUCUGUCGUCAAAACGACAUCUGUUUGUGUUUCCAGAAACGACCGCGCUCAUCGUUACCAACGGUAGAUGGCGCCAGUGCAUCAUUAAGGUCCUUUUCC-------------- ..(((((((((..(((...(((((....(((...(((((....)))))...)))....)))))....)))..))))))))).........................-------------- ( -30.70) >DroYak_CAF1 89900 117 + 1 CAGCGCCAUCUAUCGUGAGAACGACACCUGUAUGUGUUUCCAGUAACCACGGCUUC---CUCUACCGACGGUAGAUGGCGCCAGUGCAUAAUUAGGGUCCUUUUCUAAUUUCAAUGCAGA ..((((((((((((((......(((((......)))))...........(((....---.....)))))))))))))))))...(((((((((((((.....))))))))...))))).. ( -39.90) >consensus CAGCGCCAUCUAUCGUCAAAACGACAUCUGCCUGUGUUUCCAGGAACGACCGCGCUCAUCGUUACCAACGGUAGAUGGCGCCAGUGCAACAUUAAGGUCCUUUUCC______________ ..((((((((((((((...(((((.....((((((((((....)))))..)))))...)))))....))))))))))))))....................................... (-24.99 = -27.42 + 2.44)


| Location | 16,414,262 – 16,414,382 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
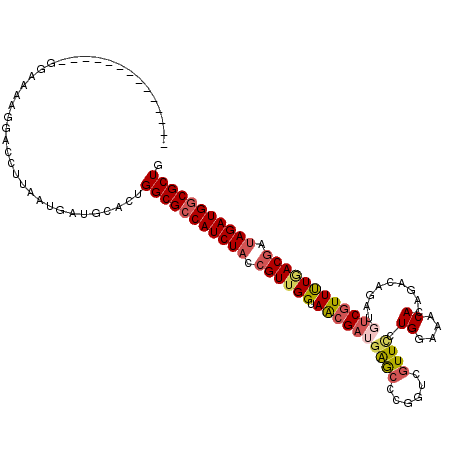
| Reading direction | reverse |
| Mean pairwise identity | 73.51 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -26.98 |
| Energy contribution | -28.47 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16414262 120 - 20766785 UAUGAUUUGAAAUUGGAAAAGGUCCUUAAUGUUGCACUGGCGCCAUCUACCGUUGGUAACGAUGAGCCCGGCCGUUCCUGGAAACACAGGCAGAUCUCGUUUUGACGAUAGAUGUCGCUG ..............(((.....))).............((((.((((((.(((..(.(((((.((.....(((.....((....))..)))...))))))))..))).)))))).)))). ( -34.50) >DroSec_CAF1 90183 99 - 1 --------------UGAGA-------UGAUGUCGCACUGGCGCCAUCUACAGUUGGUAACGAUGAGCCCGGACGUUCCUGGAAACACAGGUAGAUGUCGUUUUAACGAUAGAUGGCGCUG --------------.....-------............(((((((((((..(((((.(((((((.(((.((.....))((....))..)))...))))))))))))..))))))))))). ( -35.70) >DroSim_CAF1 85906 106 - 1 --------------GGAAAAGGACCUUAAUGAUGCACUGGCGCCAUCUACCGUUGGUAACGAUGAGCGCGGUCGUUUCUGGAAACACAAACAGAUGUCGUUUUGACGACAGAUGGCGCUG --------------........................((((((((((..(((..(.((((((.......(((((((.((....)).)))).))))))))))..)))..)))))))))). ( -36.21) >DroYak_CAF1 89900 117 - 1 UCUGCAUUGAAAUUAGAAAAGGACCCUAAUUAUGCACUGGCGCCAUCUACCGUCGGUAGAG---GAAGCCGUGGUUACUGGAAACACAUACAGGUGUCGUUCUCACGAUAGAUGGCGCUG ..(((((...((((((.........)))))))))))..((((((((((...((((..((((---((.(((...((...((....))...)).))).)).))))..)))))))))))))). ( -40.70) >consensus ______________GGAAAAGGACCUUAAUGAUGCACUGGCGCCAUCUACCGUUGGUAACGAUGAGCCCGGUCGUUCCUGGAAACACAGACAGAUGUCGUUUUGACGAUAGAUGGCGCUG ......................................(((((((((((.((((((.((((((((((......)))).((....)).........)))))))))))).))))))))))). (-26.98 = -28.47 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:14 2006