| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,389,922 – 16,390,040 |
| Length | 118 |
| Max. P | 0.522411 |

| Location | 16,389,922 – 16,390,040 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -21.50 |
| Energy contribution | -23.33 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
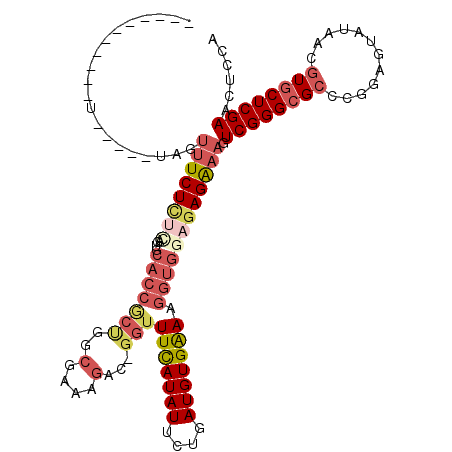
>2R_DroMel_CAF1 16389922 118 - 20766785 GAAUGCCAUACUGGCAUUAGUUUCUCUCAGUGCACCGCUGGCGAAAGAC-GGUUUCAUAUUCUGAUGUGAAAGGUGGAUAGAAAGUCGGGCGCCCGGAGUAUAACGUGCUCGAACUCCA .((((((.....))))))(((((((.((.((.((((((((.(....).)-)))(((((((....))))))).)))).)).)).)).(((....)))((((((...)))))))))))... ( -37.20) >DroVir_CAF1 99784 91 - 1 -----------AAAAAGUGG--UCUU-UAGU--------------GUGUGGCCUUUAUAUUCGGAUGUGGAACGUGGAGAGAAAGUCGGGGGCGCGCAGUAUGACAUGCUCGAACUCCA -----------....(((((--((..-((.(--------------(((((.(((..((.(((..(((.....))).....))).))..))).)))))).)).))).))))......... ( -22.30) >DroSec_CAF1 59936 99 - 1 -------------------GUUUCUCUCAGUGCACCGCUGGCGAAAGAC-GGUUUCAUAUUCUGAUGUGAAAGGUGGAGAGAAAGUCGGGCGCUCGGAGUAUAACGUGCUCGAACUCCA -------------------.((((((((....((((((((.(....).)-)))(((((((....))))))).)))))))))))).((((((((............))))))))...... ( -35.30) >DroSim_CAF1 61759 104 - 1 GAA--------------UAGUUUCUCUCAGUACACCGCUGGCGAAAGAC-GGUUUCAUAUUCUGAUGUGAAAGGUGGAGAGGAAGUCGGGCGCCCGGAGUAUAACGUGCUCGAACUCCA ...--------------...((((((((....((((((((.(....).)-)))(((((((....))))))).)))))))))))).((((((((............))))))))...... ( -34.30) >DroEre_CAF1 65293 117 - 1 GC-CGCCCUACUGGCGAUAUUUUCUCAGAGUGCACCACUGGCGAAAGAC-GGUUUCAUAUUCUGAUGUGAAAGGUGGAGAGGAAGUCGGGCGCCCGGAGUAUGACGUGCUCGAACUCCA ((-((((.....)))(((.(((((((......((((((((.(....).)-)))(((((((....))))))).))))))))))).))).)))....(((((.(((.....))).))))). ( -41.10) >DroYak_CAF1 64763 114 - 1 ----AACCUACUAAAGAUAUUUUCUCUAAGUGCACCACCGGCGAAAGAC-GGUUUCAUAUUCUGAUGUGAAAGGUGGAGAGGAAGUCGGGCGCACGGAGUAUAACGUGCUCGAACUCCA ----....((((..(((.....)))...))))((((((((.(....).)-)))(((((((....))))))).))))....(((..((((((((............))))))))..))). ( -36.10) >consensus ___________U_____UAGUUUCUCUCAGUGCACCGCUGGCGAAAGAC_GGUUUCAUAUUCUGAUGUGAAAGGUGGAGAGAAAGUCGGGCGCCCGGAGUAUAACGUGCUCGAACUCCA ....................((((((((....(((((((..(....)...)))(((((((....))))))).)))))))))))).((((((((............))))))))...... (-21.50 = -23.33 + 1.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:03 2006