
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,385,726 – 16,385,824 |
| Length | 98 |
| Max. P | 0.648137 |

| Location | 16,385,726 – 16,385,824 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.00 |
| Mean single sequence MFE | -20.15 |
| Consensus MFE | -12.82 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648137 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16385726 98 + 20766785 ACAACAAAACGGAAAUAUUUCGCUUACCAAAAAAUUGUGUGCUGCAAAAAAUUUUGAAUUUGCAAUUGCUGGUAAAGGUAAGC--------------CAACCAA----UCAAACUG ..........((...........((((((...(((((((.....((((....))))....)))))))..)))))).((....)--------------)..))..----........ ( -15.70) >DroVir_CAF1 93411 113 + 1 UCAACAAAGCGGAAGUAUUUCACUUACCAAAAAAUUGUGUGCUGCAAAAAAUUUUGCAUUUGCAAUUGCUGGUAAAGGUAAGUUGCAACCAGACCAAGAACAAGA--AGCAAAA-G ........((....)).((((..((((((...(((((((...((((((....))))))..)))))))..)))))).(((..((....))...)))........))--)).....-. ( -22.40) >DroPse_CAF1 88485 96 + 1 UCAACAAAGCGGAAAUAUUUCGCUUACCAAAAAAUUGUGUGUUGCAAAAAAUUUUGAAUUUGCAAUUGCUGGUAAAGGUAAGC--------------UAGCCCCCGC-UC-----G .......(((((...........((((((...(((((((.(((.((((....))))))).)))))))..)))))).(((....--------------..))).))))-).-----. ( -22.40) >DroGri_CAF1 69916 99 + 1 UCAACAAAGCGGAAAUAUUUCACUUACCAAAAAACUGUGUGCUUCAGAAAAUUUUGCAUUUGCAAUUGCUGGUAAAGGUAAGU--------------GAACAAAC--AUUAAAA-G ..................((((((((((......(((.......))).....(((((....((....))..))))))))))))--------------))).....--.......-. ( -20.50) >DroMoj_CAF1 108611 97 + 1 UCAGCCAGGCGGAAAUAUUUCACUUACCCAAAAAUUGUGUUUUGCAAAAAAUUUUGCAUUUGCAAUUGUUGGUCAAGGUAGGU----------------AAAAGG--UGUAAAA-G ...(((.....(((....)))((((((((((.(((((((...((((((....))))))..))))))).))).....)))))))----------------....))--)......-. ( -21.80) >DroAna_CAF1 55151 90 + 1 UCAACAAAGCGGAAAUAUUUCGCUUACCAAAAAAUUGUGUGCUGCAAAAAAUUUUGAAUUUGCAAUUGCUGGUAAAGGUAAGC--------------CAACUA------------G ........(((((.....)))))((((((...(((((((.....((((....))))....)))))))..)))))).((....)--------------).....------------. ( -18.10) >consensus UCAACAAAGCGGAAAUAUUUCACUUACCAAAAAAUUGUGUGCUGCAAAAAAUUUUGAAUUUGCAAUUGCUGGUAAAGGUAAGC______________CAACAAA____UCAAAA_G .....................(((((((..((((((.((.....))...))))))...(((((........))))))))))))................................. (-12.82 = -12.52 + -0.31)

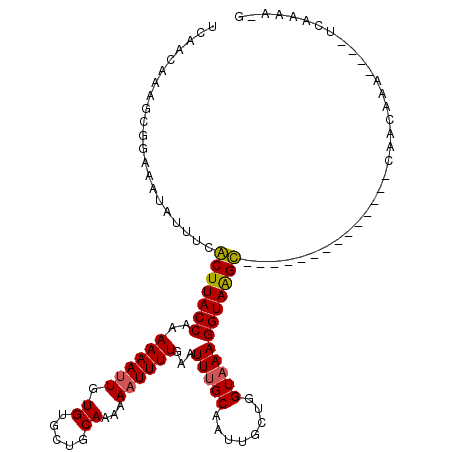
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:35:01 2006