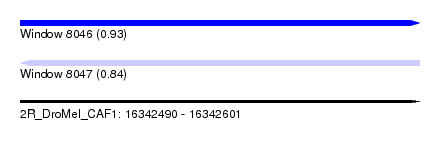
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,342,490 – 16,342,601 |
| Length | 111 |
| Max. P | 0.925741 |

| Location | 16,342,490 – 16,342,601 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.08 |
| Consensus MFE | -18.38 |
| Energy contribution | -19.75 |
| Covariance contribution | 1.38 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
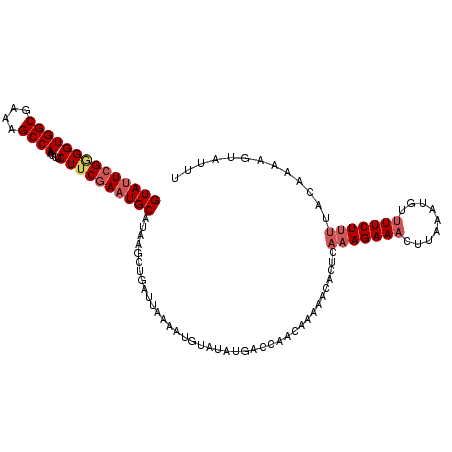
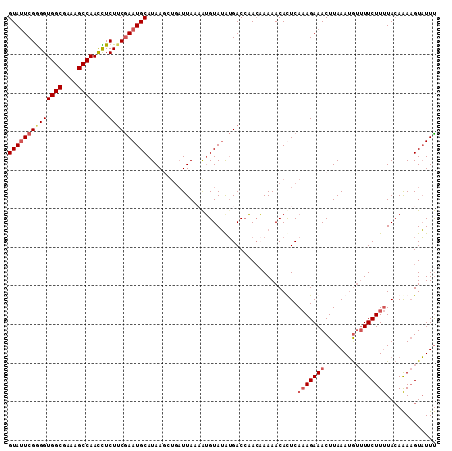
>2R_DroMel_CAF1 16342490 111 + 20766785 GUAUUCGAGGUGGCGAAAGCCAACUUCUUCAAAUGCAUAAGCUGAUUAAAAUGUAAGAGACCAACCAAAACAUUCAAAGAAACUUAAAAAUCUUCUUUUAGAAAUGUUUUU (((((.((((((((....))))....)))).)))))..............................(((((((((((((((...........))))))..).)))))))). ( -20.80) >DroSec_CAF1 12448 106 + 1 GUAUUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAA-----UUAACAUGUAUAUGACCAACAAAAACACUCAAAGAAACUUAAAUGUUUUCUUUUACAAAAGUAUUU ((((((((((((((....))).))))...)))))))....-----......((((..((.....)).........((((((((......).)))))))))))......... ( -23.60) >DroSim_CAF1 13824 111 + 1 GUAGUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAUAUGAUUAACAUGUAUAUGACCAACAAAAACACUCAAAGAAACUUAAAUGUUUUCUUUUACAAAAGUAUUU ((((((((((((((....))).))))).....((((((..((....))..))))))..)))..............((((((((......).)))))))))).......... ( -24.30) >DroYak_CAF1 16269 111 + 1 GUAUUCGGGGUGGCGAAAGCCAAUUUCUGCGAAUGCAUAAGCUGAUUAAAGGGUAUAUGACAUAGCAAAACAUUCACAGAAACUUAAAUGGUUUCUUCUACAGAAGUAUUC (((((((.((((((....))))....)).)))))))(((..(((.......(.(((.....))).)...........(((((((.....)))))))....)))...))).. ( -27.60) >consensus GUAUUCGGGGUGGCGAAAGCCAACCUCUUCGAAUGCAUAAGCUGAUUAAAAUGUAUAUGACCAACAAAAACACUCAAAGAAACUUAAAUGUUUUCUUUUACAAAAGUAUUU ((((((((((((((....))))....)))))))))).......................................(((((((.........)))))))............. (-18.38 = -19.75 + 1.38)

| Location | 16,342,490 – 16,342,601 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.93 |
| Mean single sequence MFE | -24.99 |
| Consensus MFE | -18.03 |
| Energy contribution | -16.66 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
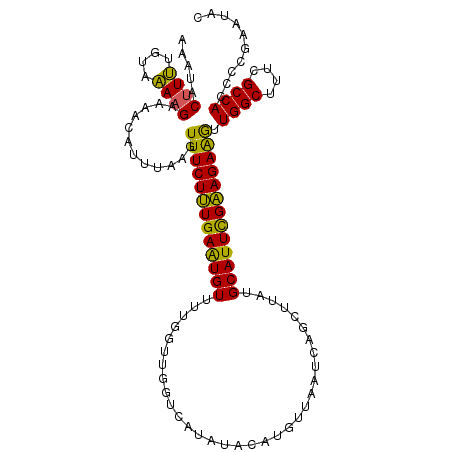
>2R_DroMel_CAF1 16342490 111 - 20766785 AAAAACAUUUCUAAAAGAAGAUUUUUAAGUUUCUUUGAAUGUUUUGGUUGGUCUCUUACAUUUUAAUCAGCUUAUGCAUUUGAAGAAGUUGGCUUUCGCCACCUCGAAUAC .......((((.....))))...((..((((((((..(((((...(((((((.............)))))))...)))))..)))))).((((....)))).))..))... ( -23.82) >DroSec_CAF1 12448 106 - 1 AAAUACUUUUGUAAAAGAAAACAUUUAAGUUUCUUUGAGUGUUUUUGUUGGUCAUAUACAUGUUAA-----UUAUGCAUUCGAAGAGGUUGGCUUUCGCCACCCCGAAUAC ....((((.(((........)))...)))).(((((((((((....(((((.(((....)))))))-----)...)))))))))))((.((((....)))).))....... ( -24.60) >DroSim_CAF1 13824 111 - 1 AAAUACUUUUGUAAAAGAAAACAUUUAAGUUUCUUUGAGUGUUUUUGUUGGUCAUAUACAUGUUAAUCAUAUUAUGCAUUCGAAGAGGUUGGCUUUCGCCACCCCGACUAC ...(((....)))...(((((((((((((....)))))))))))))(((((.((((...(((.....)))..))))..........(((.(((....)))))))))))... ( -23.90) >DroYak_CAF1 16269 111 - 1 GAAUACUUCUGUAGAAGAAACCAUUUAAGUUUCUGUGAAUGUUUUGCUAUGUCAUAUACCCUUUAAUCAGCUUAUGCAUUCGCAGAAAUUGGCUUUCGCCACCCCGAAUAC .....((((....))))..........(((((((((((((((...(((((...............)).)))....)))))))))))))))(((....)))........... ( -27.66) >consensus AAAUACUUUUGUAAAAGAAAACAUUUAAGUUUCUUUGAAUGUUUUGGUUGGUCAUAUACAUGUUAAUCAGCUUAUGCAUUCGAAGAAGUUGGCUUUCGCCACCCCGAAUAC .....((((....))))............(((((((((((((.................................))))))))))))).((((....)))).......... (-18.03 = -16.66 + -1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:50 2006