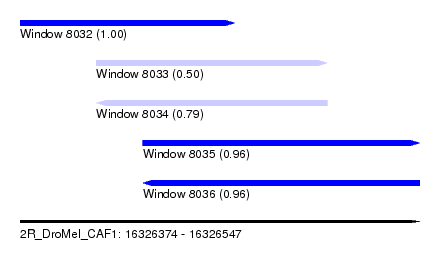
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,326,374 – 16,326,547 |
| Length | 173 |
| Max. P | 0.998930 |

| Location | 16,326,374 – 16,326,467 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 97.54 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -33.40 |
| Energy contribution | -33.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
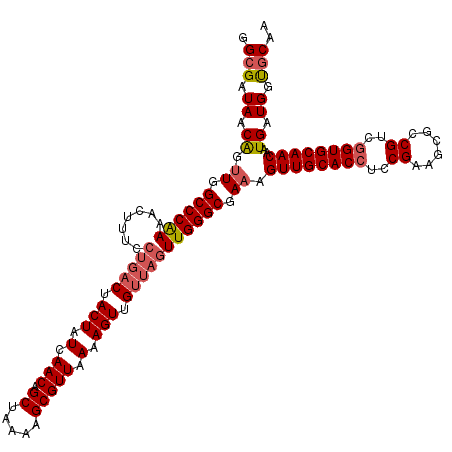
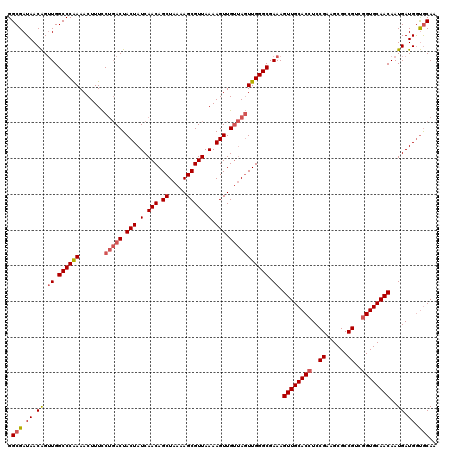
>2R_DroMel_CAF1 16326374 93 + 20766785 -GGGUAACUUGGCUUCGUUGUGUUGCAAUUGUUGUGACACCCUGAUUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGC -(((((((.((((.(((..((((..((.....))..))))..)))..))))..))))).)).........(((((..(((....)))..))))) ( -35.60) >DroSec_CAF1 29553 94 + 1 AGGGUAACUUGGCUUCGUUGUGUUGCAAUUGUUGUGACAUCCUGAUUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGC .(((((((.((((.(((..((((..((.....))..))))..)))..))))..))))).)).........(((((..(((....)))..))))) ( -34.60) >DroSim_CAF1 30038 93 + 1 -GGGUAACUUGGCUUCGUUGUGUUGCAAUUGUUGUGACACCCUGAUUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGC -(((((((.((((.(((..((((..((.....))..))))..)))..))))..))))).)).........(((((..(((....)))..))))) ( -35.60) >DroEre_CAF1 28752 93 + 1 -GGGUAACUUGGCUUCGUUGUGCUGCAAUUGUUGUGACACCCUGAUUGCCGUUGUUGCAUCAUCAUUGUUGCACCGGCGGCGCUUCGGAGGUGC -.........(((........))).............((((((((.((((((((.((((.((....)).)))).))))))))..)))).)))). ( -31.40) >DroYak_CAF1 29942 93 + 1 -GGGUAACUUGGCUUCGUUGUGUUGCAAUUGUUGUGACACCCUGAUUGCCGUUGUUGCACCAUCAUUGUUGCACCGUCGGCGCUUCGGAGGUGC -(((((((.((((.(((..((((..((.....))..))))..)))..))))..))))).)).........(((((..(((....)))..))))) ( -35.20) >consensus _GGGUAACUUGGCUUCGUUGUGUUGCAAUUGUUGUGACACCCUGAUUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGC .(((((((.((((.(((..((((((((.....))))))))..)))..))))..))))).)).........(((((..(((....)))..))))) (-33.40 = -33.28 + -0.12)


| Location | 16,326,407 – 16,326,507 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.79 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16326407 100 + 20766785 --------UGACAC----------CCUGA--UUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU --------((.(((----------(((((--.((((((((.((((.((....)).)))).))))))))..)))).))))))...........................((.....))... ( -31.60) >DroSec_CAF1 29587 100 + 1 --------UGACAU----------CCUGA--UUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU --------((.(((----------(((((--.((((((((.((((.((....)).)))).))))))))..)))).))))))...........................((.....))... ( -29.00) >DroSim_CAF1 30071 100 + 1 --------UGACAC----------CCUGA--UUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU --------((.(((----------(((((--.((((((((.((((.((....)).)))).))))))))..)))).))))))...........................((.....))... ( -31.60) >DroEre_CAF1 28785 100 + 1 --------UGACAC----------CCUGA--UUGCCGUUGUUGCAUCAUCAUUGUUGCACCGGCGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU --------((.(((----------(((((--.((((((((.((((.((....)).)))).))))))))..)))).))))))...........................((.....))... ( -31.20) >DroYak_CAF1 29975 100 + 1 --------UGACAC----------CCUGA--UUGCCGUUGUUGCACCAUCAUUGUUGCACCGUCGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU --------((.(((----------(((((--.(((((.((.((((.((....)).)))).)).)))))..)))).))))))...........................((.....))... ( -27.00) >DroAna_CAF1 30419 120 + 1 GGUGCUCCUGGCACUCCUGGUACUCCUGCUGCUGCCACUGUUGUGCCAUCAUUGUUGCACUGGCGGCGCUUCGGAGGUGCAACUUUCGCCCAAGUUGCAACUUUUAACGCUUUUAGCUGU ...(((...(((.((((.((....)).((.(((((((.(((.(((....)))....))).)))))))))...)))).((((((((......)))))))).........)))...)))... ( -39.60) >consensus ________UGACAC__________CCUGA__UUGCCGUUGUUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGU .........................((((...((((((((.((((.((....)).)))).))))))))..)))).((.((.......)))).................((.....))... (-24.94 = -24.75 + -0.19)

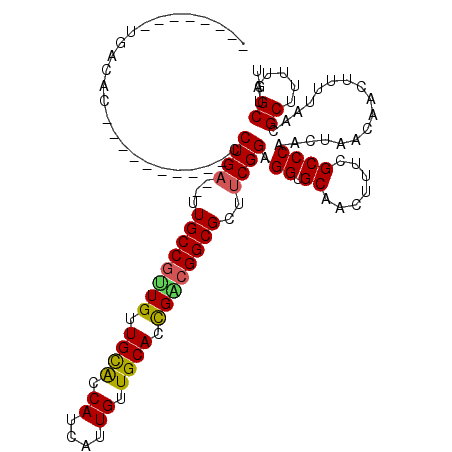
| Location | 16,326,407 – 16,326,507 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -27.94 |
| Energy contribution | -28.97 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16326407 100 - 20766785 ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAACAACGGCAA--UCAGG----------GUGUCA-------- .(((((((.(((.......))).)))))))(((....)))((((((..((...(((((.(.((((.((....)).)))).).)))))..--)).))----------))))..-------- ( -33.50) >DroSec_CAF1 29587 100 - 1 ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAACAACGGCAA--UCAGG----------AUGUCA-------- .(((((((.(((.......))).))))))).......((((((((((.(..(((((...)))))..)...)).))))))))...((((.--.....----------.)))).-------- ( -32.00) >DroSim_CAF1 30071 100 - 1 ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAACAACGGCAA--UCAGG----------GUGUCA-------- .(((((((.(((.......))).)))))))(((....)))((((((..((...(((((.(.((((.((....)).)))).).)))))..--)).))----------))))..-------- ( -33.50) >DroEre_CAF1 28785 100 - 1 ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGCCGGUGCAACAAUGAUGAUGCAACAACGGCAA--UCAGG----------GUGUCA-------- .(((((((.(((.......))).)))))))(((....)))((((((..((...((((....((((.((....)).))))....))))..--)).))----------))))..-------- ( -31.60) >DroYak_CAF1 29975 100 - 1 ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGACGGUGCAACAAUGAUGGUGCAACAACGGCAA--UCAGG----------GUGUCA-------- .(((((((.(((.......))).))))))).......((((((((((.(..(((((...)))))..)...)).))))))))...((((.--.....----------.)))).-------- ( -32.00) >DroAna_CAF1 30419 120 - 1 ACAGCUAAAAGCGUUAAAAGUUGCAACUUGGGCGAAAGUUGCACCUCCGAAGCGCCGCCAGUGCAACAAUGAUGGCACAACAGUGGCAGCAGCAGGAGUACCAGGAGUGCCAGGAGCACC ...(((....(((........)))..(((((((....)))((((.(((...(((((((..((((..........))))....))))).))....((....)).))))))))))))))... ( -36.80) >consensus ACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAACAACGGCAA__UCAGG__________GUGUCA________ .(((((((.(((.......))).)))))))((((...((((((((..((......))..))))))))..(((...(((.......)))...))).............))))......... (-27.94 = -28.97 + 1.03)

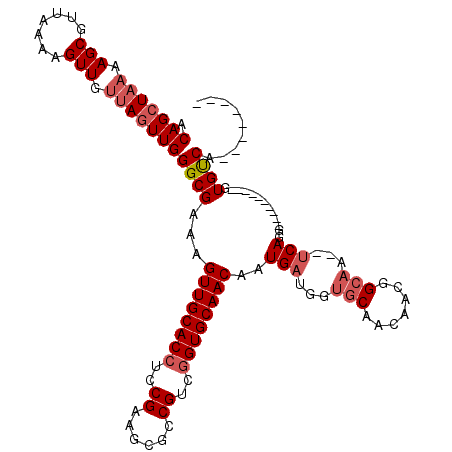
| Location | 16,326,427 – 16,326,547 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.78 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -33.69 |
| Energy contribution | -33.78 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16326427 120 + 20766785 UUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). ( -36.01) >DroSec_CAF1 29607 120 + 1 UUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). ( -36.01) >DroSim_CAF1 30091 120 + 1 UUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). ( -36.01) >DroEre_CAF1 28805 120 + 1 UUGCAUCAUCAUUGUUGCACCGGCGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). ( -34.31) >DroYak_CAF1 29995 120 + 1 UUGCACCAUCAUUGUUGCACCGUCGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). ( -35.61) >DroAna_CAF1 30459 119 + 1 UUGUGCCAUCAUUGUUGCACUGGCGGCGCUUCGGAGGUGCAACUUUCGCCCAAGUUGCAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUGCGAAAAGUUUCGGGCCA-CCGUUAUCGCC ....((((.((....))...))))((((...(((.((((((((((......)))))))(((((((..((((.(((........))).))))..)))))))....))).-)))....)))) ( -35.90) >consensus UUGCACCAUCAUUGUUGCACCGACGGCGCUUCGGAGGUGCAACUUUCGCCCAACUAACAACUUUUAACGCUUUUAGCUGUUGAUAGUAGUCAGGAAAGUUUUGGGCCAACUGUUAUCGCC ..((.........((((((((..(((....)))..))))))))....((((((((.((.(((.(((((((.....)).))))).))).)).)).......))))))...........)). (-33.69 = -33.78 + 0.09)

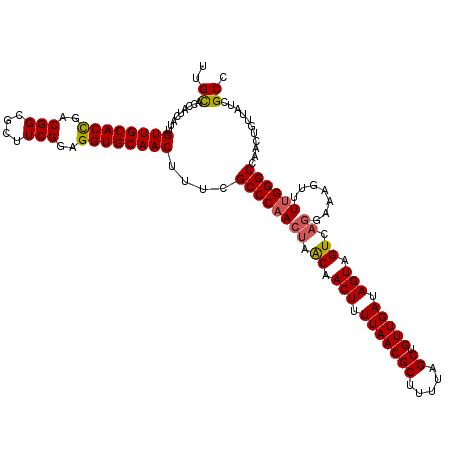
| Location | 16,326,427 – 16,326,547 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.78 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -32.59 |
| Energy contribution | -33.34 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.962063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16326427 120 - 20766785 GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAA .((......(((..(............)..)))(((((((.(((((((.(((.......))).))))))).......((((((((..((......))..))))))))..))))))))).. ( -37.20) >DroSec_CAF1 29607 120 - 1 GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAA .((......(((..(............)..)))(((((((.(((((((.(((.......))).))))))).......((((((((..((......))..))))))))..))))))))).. ( -37.20) >DroSim_CAF1 30091 120 - 1 GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAA .((......(((..(............)..)))(((((((.(((((((.(((.......))).))))))).......((((((((..((......))..))))))))..))))))))).. ( -37.20) >DroEre_CAF1 28805 120 - 1 GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGCCGGUGCAACAAUGAUGAUGCAA .(((.((.((.((.((((((.......(((((.(((.(.(((.((.....))))).).))).))))))))))).)).((((((((..((......))..))))))))..)).)).))).. ( -36.11) >DroYak_CAF1 29995 120 - 1 GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGACGGUGCAACAAUGAUGGUGCAA .((......(((..(............)..)))(((((((.(((((((.(((.......))).))))))).......((((((((..((......))..))))))))..))))))))).. ( -36.30) >DroAna_CAF1 30459 119 - 1 GGCGAUAACGG-UGGCCCGAAACUUUUCGCACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGCAACUUGGGCGAAAGUUGCACCUCCGAAGCGCCGCCAGUGCAACAAUGAUGGCACAA ((((.....((-(((..((((....))))..)))))..............(((((...((.((((((((......)))))))).))....))))))))).((((..........)))).. ( -35.90) >consensus GGCGAUAACAGUUGGCCCAAAACUUUCCUGACUACUAUCAACAGCUAAAAGCGUUAAAAGUUGUUAGUUGGGCGAAAGUUGCACCUCCGAAGCGCCGUCGGUGCAACAAUGAUGGUGCAA .(((.((.((.((.((((((.......(((((.(((.(.(((.((.....))))).).))).))))))))))).)).((((((((..((......))..))))))))..)).)).))).. (-32.59 = -33.34 + 0.75)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:39 2006