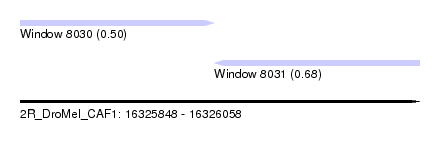
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,325,848 – 16,326,058 |
| Length | 210 |
| Max. P | 0.683768 |

| Location | 16,325,848 – 16,325,950 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -21.58 |
| Energy contribution | -22.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16325848 102 + 20766785 GUGCGUGGAAGUUUGUUGGUAUAGGUGUCGAGGA--GUCUGUGU--------GUGUUUGCU-UGGCCCGGCGGCAAAAGUCUGAUAAGCUGGUAGUUCGCAAAAAA---AAAAAA--A .((((....(((((((..(......(((((.((.--(((...((--------......)).-.)))))..))))).....)..))))))).......)))).....---......--. ( -27.20) >DroSec_CAF1 29024 109 + 1 GUGCGUGAAAGUUUGUUGGCAUAGGUGUCGGGGA--GUCUGUGU----GUGUGUGUUUGCUUUCGCCCGGCGGCAAACGUCUGAUAAGCUGGUAGUUCGCAAAAAA---AAAAAAUGA .((((....(((((((..((...(.(((((((((--((..(..(----....)..)..))))...))))))).)....).)..))))))).......)))).....---......... ( -27.50) >DroSim_CAF1 29506 114 + 1 GUGCGUGAAAGUUUGUUGGUAUAGGUGUCGGGGA--GUAUGUGU--GUGUGUGUGUUUGCUUUCGCCCGGCGGCAAAAGUCUGAUAAGCUGGUAGUUCGCAAAAAAAAAAAAAAAUGA .((((....(((((((..(....(.(((((((((--(((.(..(--......)..).)))))...))))))).)......)..))))))).......))))................. ( -28.90) >DroYak_CAF1 29407 113 + 1 GUGCGUCAAAGUUUGUUGGUAUAGGUGUCGGAGAGAGUCGGUGUAUAAGUGUGUGUUUGCUU-GAGCCGGCGGCAAAAGUCUGAUAAGCUGGUAGUUCGCACAAAA---AAAAGAA-A (((((....(((((((..(....(.((((((...((((..(..(((....)))..)..))))-...)))))).)......)..))))))).......)))))....---.......-. ( -34.20) >consensus GUGCGUGAAAGUUUGUUGGUAUAGGUGUCGGGGA__GUCUGUGU____GUGUGUGUUUGCUUUCGCCCGGCGGCAAAAGUCUGAUAAGCUGGUAGUUCGCAAAAAA___AAAAAAU_A .((((....(((((((..(....(.(((((((....((..(..(........)..)..)).....))))))).)......)..))))))).......))))................. (-21.58 = -22.07 + 0.50)


| Location | 16,325,950 – 16,326,058 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.98 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -18.41 |
| Energy contribution | -18.77 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16325950 108 - 20766785 UACUCG-AACAUCCCCCU-----------UUAGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUACACUCGCUUCCAUUUUCCUAUUUU ......-...........-----------...(((((.((((((((((((((.((....))((((......))))..)))))).)))...)))))......))))).............. ( -21.70) >DroSec_CAF1 29133 107 - 1 UACUCG-AACAUCCUCCU------------CAGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUGCACUCGCUUCCUUUUUCCUAUUUU ......-...........------------..((((((((((((((((((((.((....))((((......))))..)))))).)))...)))))).....))))).............. ( -25.20) >DroSim_CAF1 29620 107 - 1 UACUCG-AACAUCCUCCU------------CAGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUGCACUCGCUUCCUUUUUCCUAUUUU ......-...........------------..((((((((((((((((((((.((....))((((......))))..)))))).)))...)))))).....))))).............. ( -25.20) >DroEre_CAF1 28320 106 - 1 UACUCG-AACAUCCCCCU------------UUGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUACACCCGCUUCCCUUCCCCCUCU-C ......-...........------------..(((((.((((((((((((((.((....))((((......))))..)))))).)))...)))))......)))))............-. ( -21.50) >DroYak_CAF1 29520 107 - 1 UACUCG-AACAUCCCCUU------------CUGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUACACUCGCUUCCGUUUCCCCUUUUC ......-...........------------..(((((.((((((((((((((.((....))((((......))))..)))))).)))...)))))......))))).............. ( -21.50) >DroAna_CAF1 29856 114 - 1 UACUGGGAGCAGCCUCUUGCGCUCGUCCUGUGAAAGCCAAGGACAUGCAGGAUGUGAGAAGUGCACUUAAAUGCAACUCCUUCUAUGUUUUUCUUACAUACUCUUCUUUUUUUU------ ..(((.((((.((.....))))))(((((((....))..)))))...))).((((((((((.(((......)))(((.........)))))))))))))...............------ ( -28.60) >consensus UACUCG_AACAUCCCCCU____________CAGAAGCCAAGGACAUGCAGGAUGUGAGAACUGCACUUAAAUGCAACUCCUGCAAUGUUUUCCUUACACUCGCUUCCUUUUUCCUAUUUU ................................(((((.((((((((((((((.((......((((......)))))))))))).)))...)))))......))))).............. (-18.41 = -18.77 + 0.36)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:35 2006