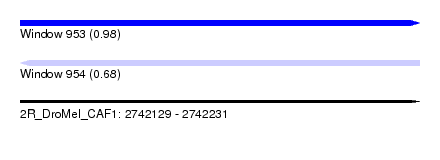
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,742,129 – 2,742,231 |
| Length | 102 |
| Max. P | 0.975797 |

| Location | 2,742,129 – 2,742,231 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -38.57 |
| Energy contribution | -38.57 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975797 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
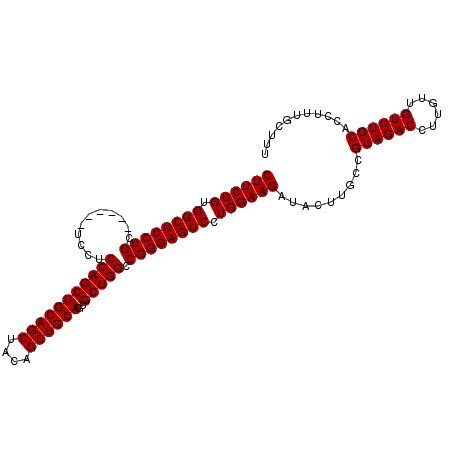
>2R_DroMel_CAF1 2742129 102 + 20766785 CUUCUGUGGCUCCUGC------UCCUCCAGCUGCAGCUACAGCUGCAGUGCUGGCCGGGAGUCCUGGAAGAUGCUUGCCGUAGACCUUGUUGUUUGCACCUUUGCUUU ((((((.((((((((.------.((....((((((((....))))))))...)).)))))))).))))))..((..(..((((((......))))))..)...))... ( -41.00) >DroSec_CAF1 26332 108 + 1 CUUCUGUGGCUCCUGCUCCAGCUCCUCCAGCUGCAGCUACAGCUGCAGUGCUGGCCGGGAGUCCUGGAAGGUACUUGCUGUAGACCUUGUUGUUUGCACCUUUGCUUU ((((((.((((((((..(((((.......((((((((....))))))))))))).)))))))).))))))(((...(.(((((((......))))))).)..)))... ( -46.51) >DroSim_CAF1 12769 102 + 1 CUUCUGUGGCUCCUGC------UCCUCCAGCUGCAGCUACAGCUGCAGUGCUGGCCGGGAGUCCUGGAAGAUACUUGCCGUAGACCUUGUUGUUUGCACCUUUGCUUU ((((((.((((((((.------.((....((((((((....))))))))...)).)))))))).)))))).........((((((......))))))........... ( -40.80) >consensus CUUCUGUGGCUCCUGC______UCCUCCAGCUGCAGCUACAGCUGCAGUGCUGGCCGGGAGUCCUGGAAGAUACUUGCCGUAGACCUUGUUGUUUGCACCUUUGCUUU ((((((.((((((((...........(((((((((((....))))))..))))).)))))))).)))))).........((((((......))))))........... (-38.57 = -38.57 + 0.00)

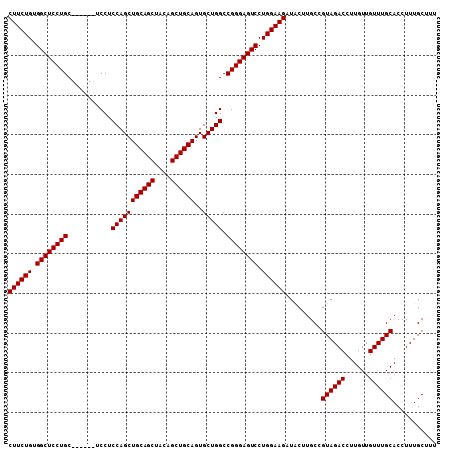
| Location | 2,742,129 – 2,742,231 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.34 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -30.77 |
| Energy contribution | -30.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
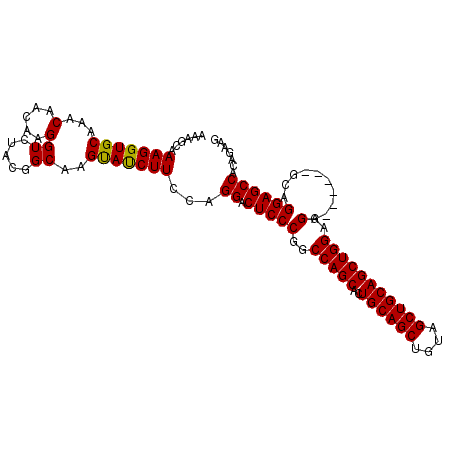
>2R_DroMel_CAF1 2742129 102 - 20766785 AAAGCAAAGGUGCAAACAACAAGGUCUACGGCAAGCAUCUUCCAGGACUCCCGGCCAGCACUGCAGCUGUAGCUGCAGCUGGAGGA------GCAGGAGCCACAGAAG ...((..((((((...(.....)((.....))..))))))(((..(.((((...(((((..((((((....))))))))))).)))------)).)))))........ ( -34.60) >DroSec_CAF1 26332 108 - 1 AAAGCAAAGGUGCAAACAACAAGGUCUACAGCAAGUACCUUCCAGGACUCCCGGCCAGCACUGCAGCUGUAGCUGCAGCUGGAGGAGCUGGAGCAGGAGCCACAGAAG ...(((....))).......(((((((......)).)))))...((.((((.(.(((((.((.(((((((....))))))).))..)))))..).))))))....... ( -40.10) >DroSim_CAF1 12769 102 - 1 AAAGCAAAGGUGCAAACAACAAGGUCUACGGCAAGUAUCUUCCAGGACUCCCGGCCAGCACUGCAGCUGUAGCUGCAGCUGGAGGA------GCAGGAGCCACAGAAG ...(((....)))...........(((..(((........(((..(.((((...(((((..((((((....))))))))))).)))------)).))))))..))).. ( -32.90) >consensus AAAGCAAAGGUGCAAACAACAAGGUCUACGGCAAGUAUCUUCCAGGACUCCCGGCCAGCACUGCAGCUGUAGCUGCAGCUGGAGGA______GCAGGAGCCACAGAAG ......(((((((...(.....)((.....))..)))))))...((.(((((..(((((..((((((....)))))))))))..)..........))))))....... (-30.77 = -30.33 + -0.44)

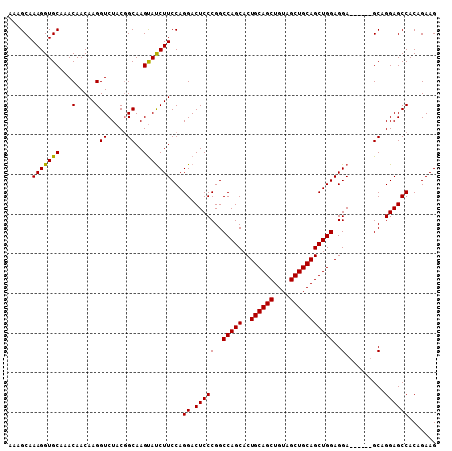
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:20 2006