
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,322,558 – 16,322,659 |
| Length | 101 |
| Max. P | 0.518953 |

| Location | 16,322,558 – 16,322,659 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.69 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -19.14 |
| Energy contribution | -18.70 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16322558 101 - 20766785 UCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA-AGUUACACAAAG-------C-----CACCGCCCACACAG-----GCCAACACUUUGUGUGUCUGUCUGCGAGUGGAAAG .....((((((((((.(((....)))...)))))..(((-((.(((((((((-------.-----....(((......)-----)).....))))))))).)).)))....)))))... ( -26.70) >DroSec_CAF1 25814 108 - 1 UCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA-AGUUACACAAAGCUUUCGGC-----CACCGCCCACACAG-----ACAAACACUUUGUGUGUCUGUCUGCGAGUGGAAAG .....(((((...((((..(((((.......((((....-))))........)))))(((-----....)))))))(((-----(((.((((.....)))).))))))...)))))... ( -30.66) >DroSim_CAF1 26235 108 - 1 UCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA-AGUUACACAAAGCUUUCGGC-----CACCGCCCACACAG-----ACAAACACUUUGUGUGUCUGUCUGCGAGUGGAAAG .....(((((...((((..(((((.......((((....-))))........)))))(((-----....)))))))(((-----(((.((((.....)))).))))))...)))))... ( -30.66) >DroEre_CAF1 25008 105 - 1 CCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA-AGUUACACAAAGUCC-GUGC-----CACCGCCCA--CAG-----GCCAACACUUUGUGUGUCUGUCUGCGAGUGGAAAG .....((((((((((.(((....)))...)))))..(((-((.((((((((((..-..((-----(........--..)-----))....)))))))))).)).)))....)))))... ( -27.80) >DroYak_CAF1 26011 106 - 1 UCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA-AGUUACACAAAGCACUUUGC-----CACCGCCCA--CAG-----GCCAACACUUUGUGUGUCUGUCUGUGAGUGGAAAG .....((((.(((((.(((....)))...))))))).))-...........((.....))-----..((((.((--(((-----((..((((.....))))..))))))).)))).... ( -29.10) >DroAna_CAF1 26677 115 - 1 UCGAAUUGGGGCCUGCGUUAAGAGAUUACCAAGCCGAGAAAGUUCAACAAAGCUUU--GCACUCACACUACGGA--CAGGGCUCGCUAAUACUUUGUGAGUCUGUCUGUGUCUGAGAUC (((..((((..(((......)).)....))))..))).((((((......))))))--...((((.((.(((((--((((.(((((.........)))))))))))))))).))))... ( -35.70) >consensus UCUAAUCCAUGUUUGUGUUGAAAGAUUACCAAACUGAGA_AGUUACACAAAGCUUU__GC_____CACCGCCCA__CAG_____GCCAACACUUUGUGUGUCUGUCUGCGAGUGGAAAG .....((((((((((.(((....)))...)))))..(((.((.(((((((((.......................................))))))))).)).)))....)))))... (-19.14 = -18.70 + -0.44)

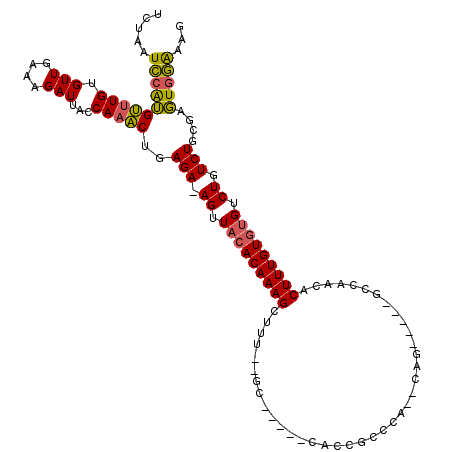
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:31 2006