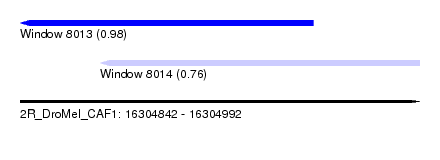
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,304,842 – 16,304,992 |
| Length | 150 |
| Max. P | 0.983184 |

| Location | 16,304,842 – 16,304,952 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -16.81 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.98 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16304842 110 - 20766785 CGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGAAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCACACAACAUCAGGCUCACACCUACACACCUCU .((((((((((((....((((...............))))((....)))))))))))((((............................))))......)))........ ( -14.75) >DroSec_CAF1 8226 110 - 1 AGUAAACAUUUUUUUAAUUUAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCACACAACAUCAGGCUCACACCUACACACCUCU .((((((((((((..............((((.....))))((....)))))))))))((((............................))))......)))........ ( -17.59) >DroSim_CAF1 7632 110 - 1 CGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAUACAACAUCAGGCUCACACCUACACACCUCU .((((((((((((..............((((.....))))((....)))))))))))((((............................))))......)))........ ( -17.69) >DroEre_CAF1 7201 105 - 1 CGUAAACAUUU-UUUAAUUCAUUCUCAUCCAAAACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCACACACCAUCACGCUUACACCUACAC----CU .((.(((((((-((.............((((.....))))((....))))))))))).))..............((((............))))..........----.. ( -17.00) >DroYak_CAF1 7911 106 - 1 CGUAAACAUUUUUUUAAUUCAUUCUCAUCCAAAACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCACACACCAUCACGCUUACACCUACAC----CU .((.(((((((((..............((((.....))))((....))))))))))).))..............((((............))))..........----.. ( -17.00) >consensus CGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCACACAACAUCAGGCUCACACCUACACACCUCU .((.(((((((((..............((((.....))))((....))))))))))).))...............(((............)))................. (-13.58 = -13.98 + 0.40)

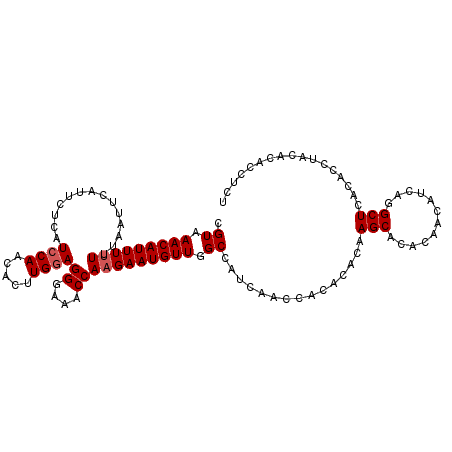
| Location | 16,304,872 – 16,304,992 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16304872 120 - 20766785 CAGCCAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGAAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAC ((((((((.....)))).))))........(.(((.((...((.(((((((((....((((...............))))((....))))))))))).))..)).))).).......... ( -25.26) >DroSec_CAF1 8256 120 - 1 CAGCCAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUAGUAAACAUUUUUUUAAUUUAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAC ((((((((.....)))).))))........(.(((.((...((.(((((((((..............((((.....))))((....))))))))))).))..)).))).).......... ( -28.20) >DroSim_CAF1 7662 120 - 1 CAGCCAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAU ((((((((.....)))).))))........(.(((.((...((.(((((((((..............((((.....))))((....))))))))))).))..)).))).).......... ( -28.20) >DroEre_CAF1 7227 119 - 1 CAGCCAUUUCACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUU-UUUAAUUCAUUCUCAUCCAAAACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAC ((((((((.....)))).))))........(.(((.((...((.(((((((-((.............((((.....))))((....))))))))))).))..)).))).).......... ( -28.20) >DroYak_CAF1 7937 120 - 1 CAGCCAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUUUUUUAAUUCAUUCUCAUCCAAAACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAC ((((((((.....)))).))))........(.(((.((...((.(((((((((..............((((.....))))((....))))))))))).))..)).))).).......... ( -28.20) >DroAna_CAF1 8557 115 - 1 CGGCUAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUUUUUUAAUUCAUUCUCCUCCAACACUAGAAGACCAGAAAAGAAUGCUGGCCAUCAACCACACACA----- .((........))..((((((........)))(((.((...((...(((((((((......((((...........))))......)))))))))...))..)).))))))....----- ( -17.70) >consensus CAGCCAUUUUACCAAUGUGCUGAAAAUCAAGCGGUCGAGUCGUAAACAUUUUUUUAAUUCAUUCUCAUCCAACACUUGGAGGGAAACCAAGAAUGUUGGCCAUCAACCACACACAAGCAC ((((((((.....)))).))))........(.(((.((...((.(((((((((..............((((.....))))((....))))))))))).))..)).))).).......... (-21.86 = -22.75 + 0.89)
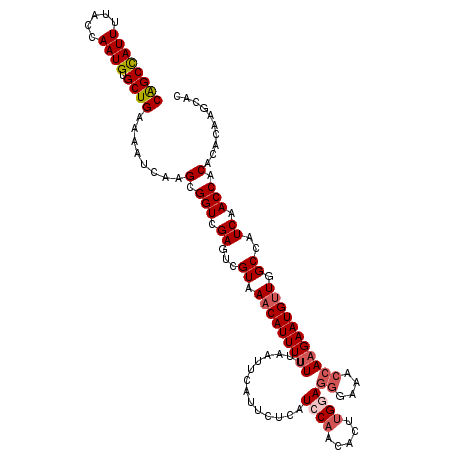
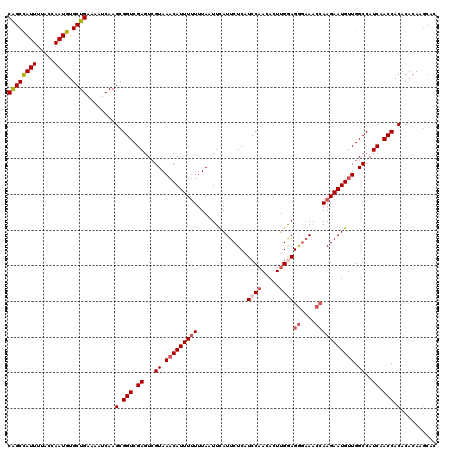
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:18 2006