| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
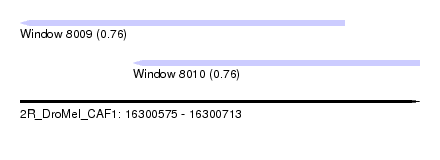
| Location | 16,300,575 – 16,300,713 |
| Length | 138 |
| Max. P | 0.764892 |

| Location | 16,300,575 – 16,300,687 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.23 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -28.72 |
| Energy contribution | -29.83 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764892 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16300575 112 - 20766785 CCUCCGAGUUCUGCUCAAGAUUCUCCUCCAGGGGAAUGGCCAUUGCCAUGGCUCCGAAGAGCGCCAGGCACAGGAAGAGUACCAGUUUGGUGCUCAUGGUGAA-GAGGGUGUU ((((.(((.....)))...(((((((....))))))).(((((((((.((((.(......).))))))))......(((((((.....))))))))))))...-))))..... ( -44.40) >DroSec_CAF1 3371 112 - 1 CCUCCGAGUUCUGCCCUAGAUUCUCCUCUAGGGGAUUGGACAUAGCCAUGGCUCCGAAGAGCGCCAGGCACAGGAAGAGUACCAGUUUGGUGCUCAUGGUGAA-GAGGGUGUU ((((....((((((((((((......)))))))...........(((.((((.(......).))))))).))))).(((((((.....)))))))........-))))..... ( -43.10) >DroSim_CAF1 3418 112 - 1 CCUCCGUGUUCUGCCCAAGAUUCUCCUCUAGGGGAUUGGCCAUGGCCAUGGCUCCGAAGAGCGCCAGGCACAGGAAGAGUACCAGUUUGGUGCUCAUGGUGAA-GAGGGUGUU ............((((....(((.((....((((..((((....))))...))))...(((((((((((...((.......)).)))))))))))..)).)))-..))))... ( -42.60) >DroEre_CAF1 2710 100 - 1 CC------------UGAAGAUUCUCCCUCAUGGGAACAGCCAUUGCCAUGGCUCCGCAGAGCGCCACGCACAGGAAGAGUACCAGUUUGGUGCUCAUGAUGGA-GAGGGUGUU ..------------........(.(((((.((....))(((((....)))))((((((((((((((.((...((.......)).)).)))))))).)).))))-))))).).. ( -36.20) >DroYak_CAF1 3399 112 - 1 ACACCGCGUUCUGUUCGAUAUUCUCCUCCAGGGGAAGGGCCAUGGCCAUGGCUCCGCAGAGCGCCAGGCACAGGAAGAGUACCAGUUUGGUGCUCAUGGUGGA-GUGGGUGUU ((((((((((((((......((((((....))))))(((((((....))))).)))))))))))....(((.....(((((((.....)))))))...)))..-...))))). ( -51.30) >DroAna_CAF1 4157 111 - 1 GCUCGGCAGUUGGAUCGACGGUAUCCACAUCAGGACGGGCCGUGGCUAGGGCUCCGAAGAGUGCCAGGCAUAGGCAGAGAACCAAUUUGGUGCUCAUGGCAGAAUAGG--GCU ((((((.((((..(.(.(((((.(((......)))...))))).).)..))))))......(((((.((....)).(((.(((.....))).))).))))).....))--)). ( -37.80) >consensus CCUCCGAGUUCUGCUCAAGAUUCUCCUCCAGGGGAAUGGCCAUGGCCAUGGCUCCGAAGAGCGCCAGGCACAGGAAGAGUACCAGUUUGGUGCUCAUGGUGAA_GAGGGUGUU ((((...................((((....))))...(((((.(((.((((.(......).))))))).......(((((((.....))))))))))))....))))..... (-28.72 = -29.83 + 1.11)

| Location | 16,300,614 – 16,300,713 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 77.82 |
| Mean single sequence MFE | -36.00 |
| Consensus MFE | -24.35 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.762517 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16300614 99 - 20766785 GCUAUUCAACU------CCUGGCUGUUCUCCACCUCCGAGUUCUGCUCAAGAUUCUCCUCCAGGGGAAUGGCCAUUGCCAUGGCUCCGAAGAGCGCCAGGCACAG ...........------((((((.(((((.(......(((.....)))...(((((((....)))))))((((((....))))))..).)))))))))))..... ( -34.20) >DroSec_CAF1 3410 99 - 1 GCUGUUCAGCU------CCUGGCUGUUCUCCACCUCCGAGUUCUGCCCUAGAUUCUCCUCUAGGGGAUUGGACAUAGCCAUGGCUCCGAAGAGCGCCAGGCACAG (((....))).------((((((.(((((.(...(((((....(.(((((((......))))))).))))))...(((....)))..).)))))))))))..... ( -36.40) >DroSim_CAF1 3457 99 - 1 GCUGUUCAGCU------CCUGGCUGUUCUCCACCUCCGUGUUCUGCCCAAGAUUCUCCUCUAGGGGAUUGGCCAUGGCCAUGGCUCCGAAGAGCGCCAGGCACAG (((....))).------((((((.(((((.(....(((((..(((.((((...(((((....))))))))).)).)..)))))....).)))))))))))..... ( -37.00) >DroEre_CAF1 2749 87 - 1 GCUGGUUAGCU------CCUGGCUGUCCUCCACC------------UGAAGAUUCUCCCUCAUGGGAACAGCCAUUGCCAUGGCUCCGCAGAGCGCCACGCACAG ((((((..(((------(.(((((((..(((.(.------------(((.(......).))).)))))))))))..((....))......)))))))).)).... ( -29.40) >DroYak_CAF1 3438 99 - 1 GCUGUUCAGCU------CCUGGCUGUUCUCCAACACCGCGUUCUGUUCGAUAUUCUCCUCCAGGGGAAGGGCCAUGGCCAUGGCUCCGCAGAGCGCCAGGCACAG (((....))).------((((((((((....))))....(((((((......((((((....))))))(((((((....))))).)))))))))))))))..... ( -40.30) >DroAna_CAF1 4195 105 - 1 GCUGUUCAGCUCCUGCUCCUGGCUGUUCUCCAGCUCGGCAGUUGGAUCGACGGUAUCCACAUCAGGACGGGCCGUGGCUAGGGCUCCGAAGAGUGCCAGGCAUAG (((.....((((..((.(((((((....(((((((....)))))))...(((((.(((......)))...))))))))))))))......))))....))).... ( -38.70) >consensus GCUGUUCAGCU______CCUGGCUGUUCUCCACCUCCGAGUUCUGCUCAAGAUUCUCCUCCAGGGGAAUGGCCAUGGCCAUGGCUCCGAAGAGCGCCAGGCACAG (((....))).......((((((.(((((.(........................((((....))))..((((((....))))))..).)))))))))))..... (-24.35 = -24.97 + 0.61)

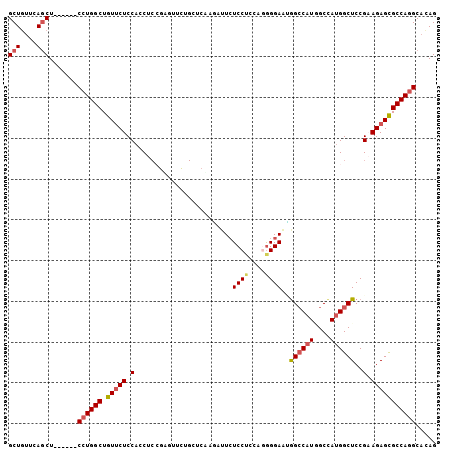
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:14 2006