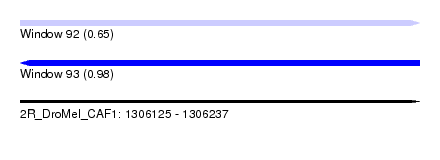
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,306,125 – 1,306,237 |
| Length | 112 |
| Max. P | 0.977377 |

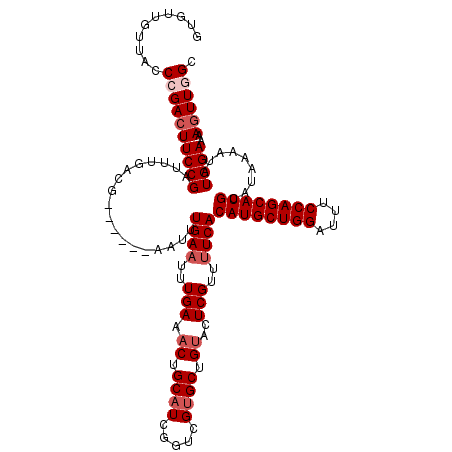
| Location | 1,306,125 – 1,306,237 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.77 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1306125 112 + 20766785 --GUUGUUGCCCGACUUCCGAUUUGACG------AAUUUGAAUUUGAAACUGCAUCGGUCGUGCUGUACUCGUUUUCACAUGCUGGAUUUCCAGCAUGCAUAAAAUAUGGAAAAGUUUGC --((((.....))))(((((((((....------....((((..(((.((.((((.....)))).))..)))..))))((((((((....))))))))....)))).)))))........ ( -28.10) >DroSec_CAF1 24506 114 + 1 GUGUUGUUACCCGACUUCCGAUUUGACG------AAUUUGAAUUUGAAACUGCAUCGGUCGUGCUGUGCUCGUUUUCACAUGCUGGAUUUCCAGCAUGCAUAAAAUAUGGAAAAGUUGGC ..........((((((((((((((....------....((((..(((.((.((((.....)))).))..)))..))))((((((((....))))))))....)))).))))..)))))). ( -34.10) >DroSim_CAF1 3497 114 + 1 GUGUUGUUACCCGACUUCCGAUUUGACG------AAUUUGAAUUUGAAACUGCAUCGGUCGUGCUGUGCUCGUUCUCACAUGCUGGAUUUCCAGCAUGCAUAAAAUAUGGAAAAGUUGGC ..........((((((((((((((....------...........(((...(((.(((.....))))))...)))...((((((((....))))))))....)))).))))..)))))). ( -31.80) >DroEre_CAF1 7285 117 + 1 GUGU---UGCCCGAAUUCCGAUUUGACGGAAUUGAAUUUGAAUUUGAAACUGCAUCGGUCGGGCUGCACUCGUUUUCACAUGCUGGAUUUCCAGCAUGCAUAAAUUAUGGAAAAGUUAGC ((((---.((((((...((((..((.(((..(..(((....)))..)..))))))))))))))).))))...(((((.((((((((....))))))))((((...)))))))))...... ( -41.10) >consensus GUGUUGUUACCCGACUUCCGAUUUGACG______AAUUUGAAUUUGAAACUGCAUCGGUCGUGCUGUACUCGUUUUCACAUGCUGGAUUUCCAGCAUGCAUAAAAUAUGGAAAAGUUGGC ..........((((((((((..................((((..(((.((.((((.....)))).))..)))..))))((((((((....)))))))).........))))..)))))). (-25.27 = -26.77 + 1.50)

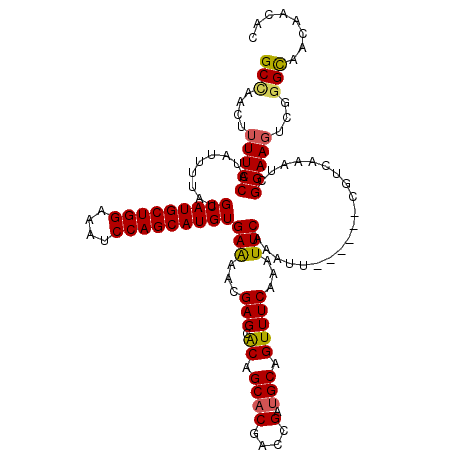
| Location | 1,306,125 – 1,306,237 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.31 |
| Mean single sequence MFE | -30.67 |
| Consensus MFE | -26.73 |
| Energy contribution | -26.72 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1306125 112 - 20766785 GCAAACUUUUCCAUAUUUUAUGCAUGCUGGAAAUCCAGCAUGUGAAAACGAGUACAGCACGACCGAUGCAGUUUCAAAUUCAAAUU------CGUCAAAUCGGAAGUCGGGCAACAAC-- ((.....(((((.........(((((((((....)))))))))....((((((((.((((....).))).))...........)))------)))......)))))....))......-- ( -27.80) >DroSec_CAF1 24506 114 - 1 GCCAACUUUUCCAUAUUUUAUGCAUGCUGGAAAUCCAGCAUGUGAAAACGAGCACAGCACGACCGAUGCAGUUUCAAAUUCAAAUU------CGUCAAAUCGGAAGUCGGGUAACAACAC (((....(((((.........(((((((((....)))))))))((...((.((...)).))...((((.(((((.......)))))------))))...)))))))...)))........ ( -29.80) >DroSim_CAF1 3497 114 - 1 GCCAACUUUUCCAUAUUUUAUGCAUGCUGGAAAUCCAGCAUGUGAGAACGAGCACAGCACGACCGAUGCAGUUUCAAAUUCAAAUU------CGUCAAAUCGGAAGUCGGGUAACAACAC (((....(((((.........(((((((((....)))))))))..((.((.((...)).))...((((.(((((.......)))))------))))...)))))))...)))........ ( -30.00) >DroEre_CAF1 7285 117 - 1 GCUAACUUUUCCAUAAUUUAUGCAUGCUGGAAAUCCAGCAUGUGAAAACGAGUGCAGCCCGACCGAUGCAGUUUCAAAUUCAAAUUCAAUUCCGUCAAAUCGGAAUUCGGGCA---ACAC (((...(((((..........(((((((((....))))))))))))))..)))...((((((..((.......)).............((((((......)))))))))))).---.... ( -35.10) >consensus GCCAACUUUUCCAUAUUUUAUGCAUGCUGGAAAUCCAGCAUGUGAAAACGAGCACAGCACGACCGAUGCAGUUUCAAAUUCAAAUU______CGUCAAAUCGGAAGUCGGGCAACAACAC (((....(((((.........(((((((((....)))))))))(((...(((.((.((((....).))).)))))...)))....................)))))...)))........ (-26.73 = -26.72 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:24:47 2006