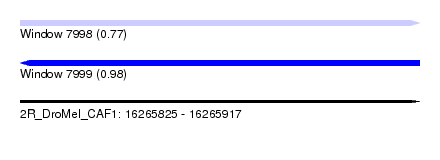
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,265,825 – 16,265,917 |
| Length | 92 |
| Max. P | 0.983465 |

| Location | 16,265,825 – 16,265,917 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 69.40 |
| Mean single sequence MFE | -22.08 |
| Consensus MFE | -11.99 |
| Energy contribution | -12.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
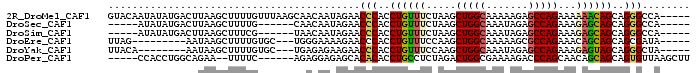
>2R_DroMel_CAF1 16265825 92 + 20766785 -----UGGCCCUGCUGUUUUUUCUGGCUCUUUUUGCCAGCUUAGAAACAGGUGGGUUCUAUUGUUGCUUAAACAAAAGCUUAAGUCAUAUAUUGUAC -----.(((((..((((((((.(((((.......)))))...))))))))..)))))....(((.((((((........)))))).)))........ ( -26.20) >DroSec_CAF1 41028 81 + 1 -----UGGCCCUGCUGCUCUUUCUGGCUCUAUUUGCCAGCUUAGAAACAGGUGGGUUCUAUUGUUG------CAAAAGCUUAAGUCAUAUAU----- -----((((((..((..(((..(((((.......)))))...)))....))..))..........(------(....))....)))).....----- ( -19.70) >DroSim_CAF1 41535 81 + 1 -----UGGCCCUGCUGCUCUUUCUGGCUCUAUUUGCCAGCUUAGAAACAGGUGGGUUCUAUUGUUA------CGAAAGCUUAAGUCAUAUAU----- -----((((((..((..(((..(((((.......)))))...)))....))..))...........------(....).....)))).....----- ( -19.70) >DroEre_CAF1 46295 80 + 1 -----UAUCGCUGCUGCUGUUUCUGGCGCUUUUUGCCAGCUUGGAAACAGGUGGGUUCUUUUCCCA---GCACAAAAGCUUAUU---------CUAA -----....(((..((((.....(((((.....)))))(((((....)))))(((.......))))---)))....))).....---------.... ( -23.50) >DroYak_CAF1 54442 81 + 1 -----UAGCCCUGCUACUCUUUCUGGCUCUAUUUGCCAGCUUGGAAACAGGUGGGUUCUUCUCUCA---GCACAAAAGCUUAUU--------UGUAA -----.(((((............((((.......))))(((((....))))))))))........(---((......)))....--------..... ( -20.50) >DroPer_CAF1 54654 84 + 1 AAGCUUAACACUGCUGCUGUUGCUGGGUCUUUUCGCCAGUCUAGAGGCAGGUGUGUGCUCUCCUCU------GAAAA--UUCUGCCAGGUGG----- .(((...((((((((.(((..((((((......).))))).))).)))).))))..)))..(((((------(....--......)))).))----- ( -22.90) >consensus _____UAGCCCUGCUGCUCUUUCUGGCUCUAUUUGCCAGCUUAGAAACAGGUGGGUUCUAUUCUCA______CAAAAGCUUAAGUCAUAUAU_____ ......(((((.(((..(((..(((((.......)))))...)))....))))))))........................................ (-11.99 = -12.18 + 0.20)

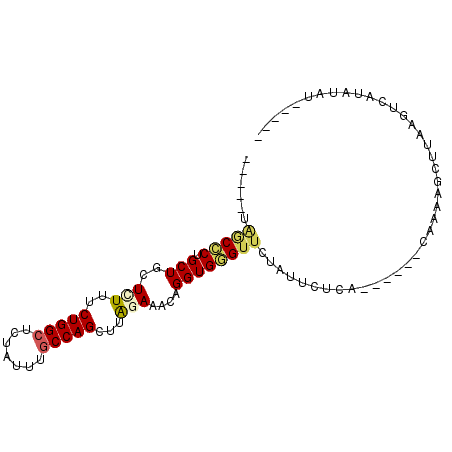
| Location | 16,265,825 – 16,265,917 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 69.40 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.33 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16265825 92 - 20766785 GUACAAUAUAUGACUUAAGCUUUUGUUUAAGCAACAAUAGAACCCACCUGUUUCUAAGCUGGCAAAAAGAGCCAGAAAAAACAGCAGGGCCA----- ..........((.(((((((....))))))))).........(((..((((((.....(((((.......)))))...))))))..)))...----- ( -24.60) >DroSec_CAF1 41028 81 - 1 -----AUAUAUGACUUAAGCUUUUG------CAACAAUAGAACCCACCUGUUUCUAAGCUGGCAAAUAGAGCCAGAAAGAGCAGCAGGGCCA----- -----.............((....)------)..............((((((.((...(((((.......)))))....)).))))))....----- ( -20.40) >DroSim_CAF1 41535 81 - 1 -----AUAUAUGACUUAAGCUUUCG------UAACAAUAGAACCCACCUGUUUCUAAGCUGGCAAAUAGAGCCAGAAAGAGCAGCAGGGCCA----- -----...(((((.........)))------)).............((((((.((...(((((.......)))))....)).))))))....----- ( -19.60) >DroEre_CAF1 46295 80 - 1 UUAG---------AAUAAGCUUUUGUGC---UGGGAAAAGAACCCACCUGUUUCCAAGCUGGCAAAAAGCGCCAGAAACAGCAGCAGCGAUA----- ....---------.....(((....(((---(((((((((.......)).)))))...(((((.......)))))...)))))..)))....----- ( -21.70) >DroYak_CAF1 54442 81 - 1 UUACA--------AAUAAGCUUUUGUGC---UGAGAGAAGAACCCACCUGUUUCCAAGCUGGCAAAUAGAGCCAGAAAGAGUAGCAGGGCUA----- .((((--------((......)))))).---...............((((((......(((((.......))))).......))))))....----- ( -19.72) >DroPer_CAF1 54654 84 - 1 -----CCACCUGGCAGAA--UUUUC------AGAGGAGAGCACACACCUGCCUCUAGACUGGCGAAAAGACCCAGCAACAGCAGCAGUGUUAAGCUU -----.(..((((.....--...))------))..).((((((((..((((.....(.((((.(......))))))....))))..))))...)))) ( -17.40) >consensus _____AUAUAUGACAUAAGCUUUUG______UAACAAUAGAACCCACCUGUUUCUAAGCUGGCAAAAAGAGCCAGAAAGAGCAGCAGGGCCA_____ ..........................................(((..((((((.....(((((.......)))))...))))))..)))........ (-14.22 = -14.33 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:03 2006