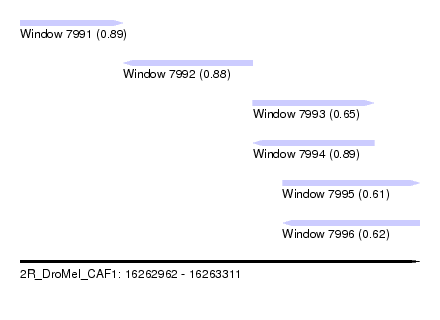
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,262,962 – 16,263,311 |
| Length | 349 |
| Max. P | 0.890530 |

| Location | 16,262,962 – 16,263,052 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.31 |
| Mean single sequence MFE | -22.48 |
| Consensus MFE | -10.78 |
| Energy contribution | -14.80 |
| Covariance contribution | 4.02 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16262962 90 + 20766785 UCAGUAUGCA-UUUCCAUUUGAGGAGCUGAGAAAUCUGUCAAGUGAAAGCUAGAAGCAAUCAAUAUCGAUUGUCAACAGAUUUUUCGCUCA ((((.(((..-....))))))).((((.((((((((((((.(((....))).)).((((((......))))))...)))))))))))))). ( -28.50) >DroSec_CAF1 38129 90 + 1 UCAAUAUGCA-UUUCCAUUUGGGGAGUUGAGCAAUCUGUCAAGUGAAAGCUAGAAGCAAUCAAUAUCGAUUGUCAACAGAUUUUUUGCUCA .........(-(((((.....))))))(((((((((((((.(((....))).)).((((((......))))))...)))))....)))))) ( -23.80) >DroSim_CAF1 38597 90 + 1 UCAAUAUGCA-UUUCCAUUUGGGGAGUUGAGGAAUCUGUCAAGUGAAAACUAGAAGCAAUCAAUAUCGAUUGUCAACAGAUUUUUUGCUCA ..........-..(((....)))((((.((((((((((((.(((....))).)).((((((......))))))...)))))))))))))). ( -23.30) >DroEre_CAF1 43390 80 + 1 -----------AUUCCAUUCGUCGAGUUGAGCAUUCUGUCAAGUGAAAGCUAAAGGCAAUCAAUAUCGAUUGUCAACAGAUUUUUUGCUCA -----------................((((((.(((((..(((....)))...(((((((......))))))).))))).....)))))) ( -22.70) >DroYak_CAF1 51512 82 + 1 UCAAUAUACUUUUUGCAUUCAUCGAGAUGAGCAAUGUGCCACGUGAAAGCUAGAAGCAAUCAAUUGCGAUUGUCAACCGAUU--------- ........(((((.((.((((.((...((.((.....)))))))))).)).)))))(((((......)))))..........--------- ( -14.10) >consensus UCAAUAUGCA_UUUCCAUUUGGGGAGUUGAGCAAUCUGUCAAGUGAAAGCUAGAAGCAAUCAAUAUCGAUUGUCAACAGAUUUUUUGCUCA .......................((((.(((.(((((((..(((....)))....((((((......))))))..)))))))))).)))). (-10.78 = -14.80 + 4.02)

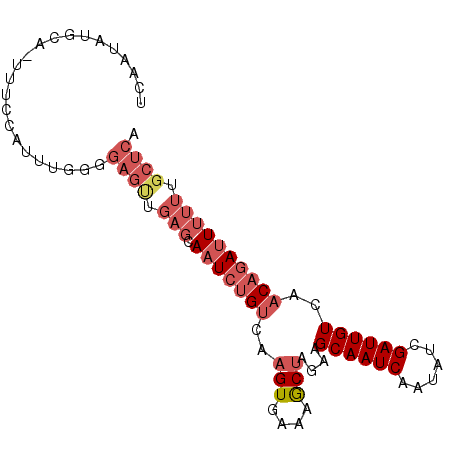
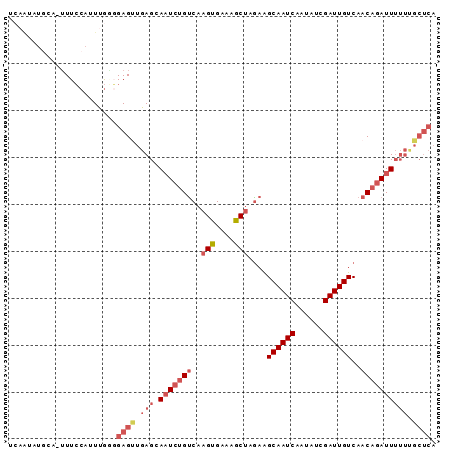
| Location | 16,263,052 – 16,263,165 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -11.78 |
| Energy contribution | -12.54 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.878972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16263052 113 - 20766785 GGCAAUUAGGGAAG-GCGGAAUAUGUCCUACAUCACAUGAUUUUGGCAUAUUCCCGUACAUUAUUCGACUUACUGACUACUUAAAGCAAAAAA---ACUUAAACUCUACUAGUUGCU ((((((((((((..-((((((((((.((...(((....)))...)))))))).))))................((.((......)))).....---........))).))))))))) ( -25.20) >DroSec_CAF1 38219 110 - 1 GGCAAUUAGGGUGG-GCGGAAUAUGUCCUACAUCACAUGAUUUUGGCAUAUUCCCGCACAUUAUUCGAGUU-----UUGCUUAAAGCAAAAAA-AAGAAGAAACUCUUCUAGGCGCC (((..((((((((.-(.((((((((.((...(((....)))...))))))))))).)))....(((...((-----((((.....))))))..-..)))........)))))..))) ( -34.00) >DroSim_CAF1 38687 111 - 1 GGCAAUUAGGGUGG-GCGGAAUAUGUCCUACAUCACAUGAUUUUGGCAUAUUCCCGCACAUUAUUCGAGUU-----UUGCUUAAAGCAAAACAAAAAACGAAACUCUUCUAGACGCC (((..((((((((.-(.((((((((.((...(((....)))...))))))))))).))).........(((-----((((.....)))))))...............)))))..))) ( -34.10) >DroEre_CAF1 43470 111 - 1 GGCAAUUAGGGAUG-GCGGAAUAUUUCCUACAACACACGUUCCUGGAAUAUUUCAGUACAUUACUCGAGUUACUGG-AGCUUAAAGCAAAAGA----AUGCAACUCUACUGGAUGCU ((((.(((((((((-..(((.....))).........))))))))).....(((((((.....(((.((...)).)-))......(((.....----.))).....))))))))))) ( -23.50) >DroYak_CAF1 51594 91 - 1 GCCAAUUAGGGAUGUGCAGAAUAUUUCCUACAUCACACCAUCCUGGAAUC------------AUUCAAGUUACUCA-AGCCUAAAGUAUUUGC----AUACUACUCUA--------- ......(((((.((((((((((..((((................))))..------------))))....((((..-.......))))..)))----)))...)))))--------- ( -15.09) >consensus GGCAAUUAGGGAGG_GCGGAAUAUGUCCUACAUCACAUGAUUUUGGCAUAUUCCCGCACAUUAUUCGAGUUACU___UGCUUAAAGCAAAAAA____AUGAAACUCUACUAGACGCC ......(((((......((((((((.((...(((....)))...))))))))))............((((........)))).....................)))))......... (-11.78 = -12.54 + 0.76)


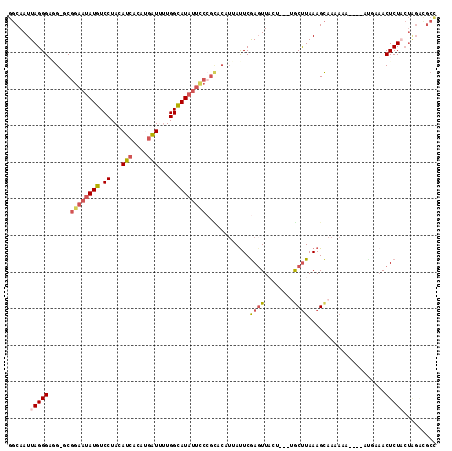
| Location | 16,263,165 – 16,263,271 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -30.00 |
| Consensus MFE | -22.13 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.652672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16263165 106 + 20766785 UUUUUG-AUUCUGCUCUCC-------------AACAGGCGAACAUCCUGCGGCCAUGCAACACAAUUCGAGUAGCAACAGCAACAGCUUCUACUGCGGCAUCUGCUGCAUGCUGCUGAGC ......-.....((((...-------------......((((.....((((....))))......))))(((((((..(((....))).....((((((....))))))))))))))))) ( -29.10) >DroPse_CAF1 51943 103 + 1 UUU--G-UUUGU-UUUUCG-------------CGCAGGCGAACACCCGGCGGCUAUGCAACAUAACUCGAGCAGCAACAGCAACAGCUUUUACUGCGGCAUCUGCUGUAUGCUGCUUAGC ...--(-(((((-(.((((-------------(....)))))......(((....))))))).)))..((((((((((((((...(((........)))...)))))).))))))))... ( -32.30) >DroGri_CAF1 19981 100 + 1 -AUCUU-AUUCU-----UU-------------UGCAGGUGAGCACCCGGCGGCCAUGCAGCACAACUCAAGCAGCAACAGCAACAGUUUCUACUGCGGCAUUUGUUGCAUGCUCCUCAGC -.....-.....-----.(-------------(((((((..((.....)).))).))))).........((((((((((((..((((....))))..))...)))))).))))....... ( -28.60) >DroWil_CAF1 82305 107 + 1 UUUCUGCUUUCUGGUUUCU-------------AACAGGCGAACACCCCGCGGCCAUGCAGCACAAUUCAAGCAGCAAUAGCAACAGUUUCUACUGCGGCAUCUGUUGCAUGUUGCUCAGC ...((((...(((......-------------..)))(((.......)))......))))..........(.((((((((((((((...((.....))...))))))).))))))))... ( -28.20) >DroMoj_CAF1 108873 102 + 1 UUUCCA-UUUCUA----CU-------------UGCAGGCGAACACCCGGCGGCCAUGCAGCACAAUUCAAGCAGCAACAGCAACAGUUUCUACUGCGGCAUUUGUUGCAUGCUGCUCAGC ......-......----..-------------.((.(.((......)).).))...((((((........(((((((..((..((((....))))..))..))))))).))))))..... ( -27.60) >DroAna_CAF1 17329 119 + 1 AUUUUG-AUUUCGCUUUCUUUUCACUUUCCCGUGCAGGCGAACACCCGGCGGCCAUGCAACACAACUCGAGCAGCAACAGCAACAGCUUCUACUGCGGCAUCUGCUGCAUGCUGCUGAGC ......-..(((((((......(((......))).)))))))....(((((((.((((............((((....(((....)))....))))(((....))))))))))))))... ( -34.20) >consensus UUUCUG_AUUCUG_UUUCU_____________UGCAGGCGAACACCCGGCGGCCAUGCAACACAACUCAAGCAGCAACAGCAACAGCUUCUACUGCGGCAUCUGCUGCAUGCUGCUCAGC .................................((((((............))).)))...........(((((((..(((....))).....((((((....))))))))))))).... (-22.13 = -21.47 + -0.66)

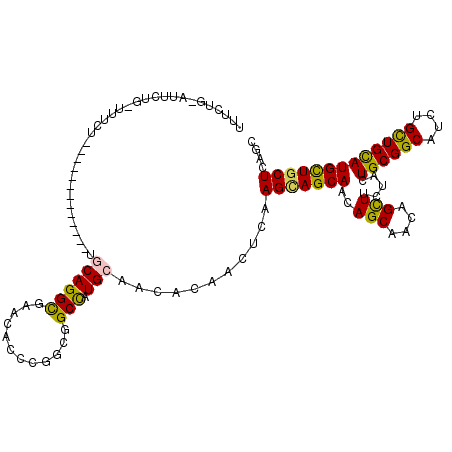
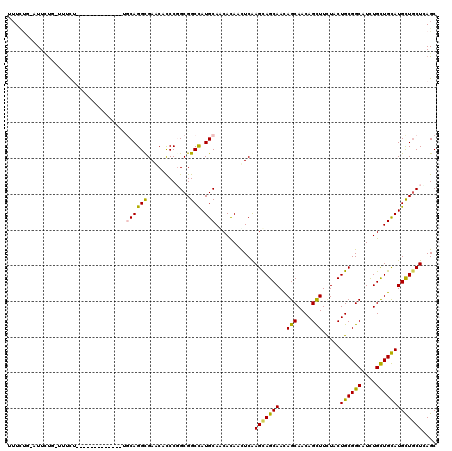
| Location | 16,263,165 – 16,263,271 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.97 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -27.87 |
| Energy contribution | -27.65 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16263165 106 - 20766785 GCUCAGCAGCAUGCAGCAGAUGCCGCAGUAGAAGCUGUUGCUGUUGCUACUCGAAUUGUGUUGCAUGGCCGCAGGAUGUUCGCCUGUU-------------GGAGAGCAGAAU-CAAAAA ((((.....(((((((((.(...((.(((((.(((....)))....)))))))...).))))))))).(((((((.......)))).)-------------)).)))).....-...... ( -39.20) >DroPse_CAF1 51943 103 - 1 GCUAAGCAGCAUACAGCAGAUGCCGCAGUAAAAGCUGUUGCUGUUGCUGCUCGAGUUAUGUUGCAUAGCCGCCGGGUGUUCGCCUGCG-------------CGAAAA-ACAAA-C--AAA ((((.(((((((((.((....)).(((((((.(((....))).)))))))....).)))))))).))))((((((((....))))).)-------------))....-.....-.--... ( -41.20) >DroGri_CAF1 19981 100 - 1 GCUGAGGAGCAUGCAACAAAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUUGAGUUGUGCUGCAUGGCCGCCGGGUGCUCACCUGCA-------------AA-----AGAAU-AAGAU- ((.(..((((..((((((...((.(((((....))))).)))))))).(((((.((.(.(((....)))))))))))))))..).)).-------------..-----.....-.....- ( -31.60) >DroWil_CAF1 82305 107 - 1 GCUGAGCAACAUGCAACAGAUGCCGCAGUAGAAACUGUUGCUAUUGCUGCUUGAAUUGUGCUGCAUGGCCGCGGGGUGUUCGCCUGUU-------------AGAAACCAGAAAGCAGAAA ...(((((.((.(((((((.(((....)))....)))))))...)).)))))...((.((((...(((..(((((.......))))).-------------.....)))...)))).)). ( -30.60) >DroMoj_CAF1 108873 102 - 1 GCUGAGCAGCAUGCAACAAAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUUGAAUUGUGCUGCAUGGCCGCCGGGUGUUCGCCUGCA-------------AG----UAGAAA-UGGAAA ((((.(((((((((((((...((.(((((....))))).))))))))..........))))))).)))).((.((((....)))))).-------------..----......-...... ( -33.60) >DroAna_CAF1 17329 119 - 1 GCUCAGCAGCAUGCAGCAGAUGCCGCAGUAGAAGCUGUUGCUGUUGCUGCUCGAGUUGUGUUGCAUGGCCGCCGGGUGUUCGCCUGCACGGGAAAGUGAAAAGAAAGCGAAAU-CAAAAU ((((.((.((((((((((.(..(.(((((((.(((....))).)))))))..)...).)))))))).)).)).)))).(((((((.(((......)))...))...)))))..-...... ( -42.10) >consensus GCUGAGCAGCAUGCAACAGAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUCGAAUUGUGCUGCAUGGCCGCCGGGUGUUCGCCUGCA_____________AGAAA_CAGAAA_CAGAAA ((((.(((((((((((((...((.(((((....))))).))))))))..........))))))).)))).((.((((....))))))................................. (-27.87 = -27.65 + -0.22)

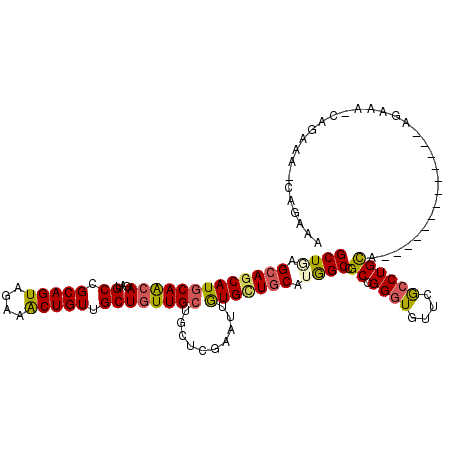
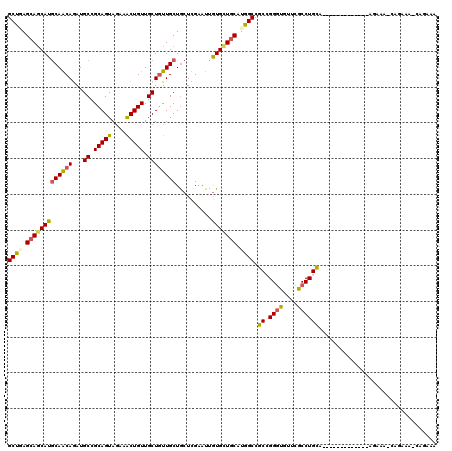
| Location | 16,263,191 – 16,263,311 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.25 |
| Mean single sequence MFE | -42.03 |
| Consensus MFE | -31.21 |
| Energy contribution | -31.27 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16263191 120 + 20766785 AACAUCCUGCGGCCAUGCAACACAAUUCGAGUAGCAACAGCAACAGCUUCUACUGCGGCAUCUGCUGCAUGCUGCUGAGCAUUGUGUCCUGCUGCCUGCAGCAUUUUUAUGAAACGGGGU ...(((((((((.((.((((((((((.(.(((((((..(((....))).....((((((....)))))))))))))..).)))))))..))))).))))..(((....)))....))))) ( -41.20) >DroVir_CAF1 92457 117 + 1 AACACCCGGCGGCCAUGCAGCACAAUUCGAGCAGCAACAGCAACAGUUUCUAUUGCGGCAUUUGUUGCAUGCUGCUCAGCAUCGUCUCCUGCUGCCUGCAGCAUUGGUAUGUGAGUG--- ..(((((((.(((.((((.(.......)((((((((...(((((((...((.....))...))))))).)))))))).)))).))).))(((((....)))))..))...)))....--- ( -40.20) >DroGri_CAF1 20001 120 + 1 AGCACCCGGCGGCCAUGCAGCACAACUCAAGCAGCAACAGCAACAGUUUCUACUGCGGCAUUUGUUGCAUGCUCCUCAGCAUCGUUUCCUGUUGCCUGCAGCACUGGUAUGUGUGUGCCG .((((.((...(((((((.(((((((..((((.((((((((..((((....))))..))...))))))(((((....))))).))))...))))..))).))).))))...)).)))).. ( -41.10) >DroWil_CAF1 82332 120 + 1 AACACCCCGCGGCCAUGCAGCACAAUUCAAGCAGCAAUAGCAACAGUUUCUACUGCGGCAUCUGUUGCAUGUUGCUCAGCAUUGUCUCCUGCUGCCUGCAGCAUUGGUUUGUCAAUGUGA ........((((....((((((.....(((((((((((((((((((...((.....))...))))))).)))))))..)).))).....)))))))))).(((((((....))))))).. ( -40.30) >DroMoj_CAF1 108895 117 + 1 AACACCCGGCGGCCAUGCAGCACAAUUCAAGCAGCAACAGCAACAGUUUCUACUGCGGCAUUUGUUGCAUGCUGCUCAGCAUUUUCUCCUGCUGCCUGCAGCAUUGGUAUGUGUGCG--- ........(((....))).(((((..(((((((((((..((..((((....))))..))..))))))).((((((.(((((........)))))...))))))))))....))))).--- ( -44.20) >DroPer_CAF1 51899 120 + 1 AACACCCGGCGGCUAUGCAACAUAACUCGAGCAGCAACAGCAACAGCUUUUAUUGCGGCAUCUGCUGUAUGCUGCUUAGCAUUGUUUCCUGCUGCCUGCAGCAUUGGUACGAGUCCGGGA ....((((((.((((((...))))....((((((((((((((...(((........)))...)))))).)))))))).)).((((..(((((((....)))))..)).))))).))))). ( -45.20) >consensus AACACCCGGCGGCCAUGCAGCACAAUUCAAGCAGCAACAGCAACAGUUUCUACUGCGGCAUCUGUUGCAUGCUGCUCAGCAUUGUCUCCUGCUGCCUGCAGCAUUGGUAUGUGAGUG_G_ ...(((..((.((...((((((........((((((...((..((((....))))..))...))))))(((((....))))).......))))))..)).))...)))............ (-31.21 = -31.27 + 0.06)

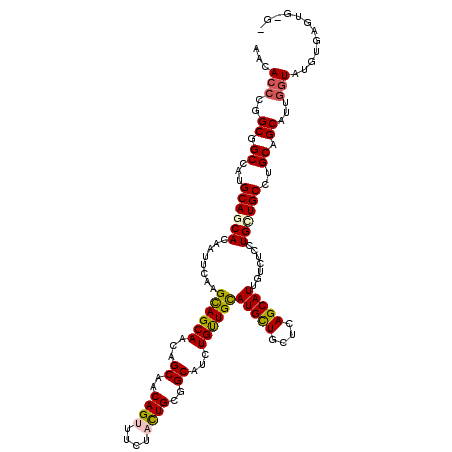
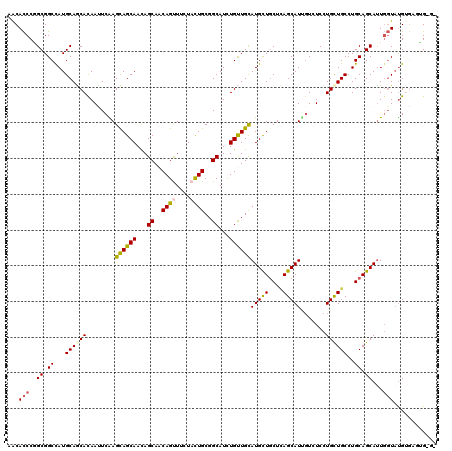
| Location | 16,263,191 – 16,263,311 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.25 |
| Mean single sequence MFE | -44.52 |
| Consensus MFE | -33.08 |
| Energy contribution | -34.25 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16263191 120 - 20766785 ACCCCGUUUCAUAAAAAUGCUGCAGGCAGCAGGACACAAUGCUCAGCAGCAUGCAGCAGAUGCCGCAGUAGAAGCUGUUGCUGUUGCUACUCGAAUUGUGUUGCAUGGCCGCAGGAUGUU ................((.((((.(((.(((..(((((((....(((((((.(((((((.(((....)))....))))))))))))))......))))))))))...))))))).))... ( -43.40) >DroVir_CAF1 92457 117 - 1 ---CACUCACAUACCAAUGCUGCAGGCAGCAGGAGACGAUGCUGAGCAGCAUGCAACAAAUGCCGCAAUAGAAACUGUUGCUGUUGCUGCUCGAAUUGUGCUGCAUGGCCGCCGGGUGUU ---.......(((((...((.((..(((((((....)(((..(((((((((.(((.....))).(((((((...)))))))...))))))))).))).))))))...)).))..))))). ( -46.80) >DroGri_CAF1 20001 120 - 1 CGGCACACACAUACCAGUGCUGCAGGCAACAGGAAACGAUGCUGAGGAGCAUGCAACAAAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUUGAGUUGUGCUGCAUGGCCGCCGGGUGCU ((((...(((......))).((((((((((.(....).........(((((.((((((...((.(((((....))))).)))))))))))))..))))).))))).....))))...... ( -44.30) >DroWil_CAF1 82332 120 - 1 UCACAUUGACAAACCAAUGCUGCAGGCAGCAGGAGACAAUGCUGAGCAACAUGCAACAGAUGCCGCAGUAGAAACUGUUGCUAUUGCUGCUUGAAUUGUGCUGCAUGGCCGCGGGGUGUU .......((((..((...((.((..((((((.((..(((.((..(((((...(((((((.(((....)))....)))))))..))))))))))..)).))))))...)).))))..)))) ( -43.10) >DroMoj_CAF1 108895 117 - 1 ---CGCACACAUACCAAUGCUGCAGGCAGCAGGAGAAAAUGCUGAGCAGCAUGCAACAAAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUUGAAUUGUGCUGCAUGGCCGCCGGGUGUU ---.(((((((...(((((((((...(((((........))))).)))))))((((((...((.(((((....))))).))))))))....))...)))).)))...(((....)))... ( -43.30) >DroPer_CAF1 51899 120 - 1 UCCCGGACUCGUACCAAUGCUGCAGGCAGCAGGAAACAAUGCUAAGCAGCAUACAGCAGAUGCCGCAAUAAAAGCUGUUGCUGUUGCUGCUCGAGUUAUGUUGCAUAGCCGCCGGGUGUU .(((((...........(((((....)))))(....)...((((.((((((((((((((..((.((((((.....)))))).))..)))))...).)))))))).))))..))))).... ( -46.20) >consensus _C_CACACACAUACCAAUGCUGCAGGCAGCAGGAGACAAUGCUGAGCAGCAUGCAACAAAUGCCGCAGUAGAAACUGUUGCUGUUGCUGCUCGAAUUGUGCUGCAUGGCCGCCGGGUGUU ..........(((((...((.((..((((((.((..(((.((..(((((((.((((((.................))))))))))))))))))..)).))))))...)).))..))))). (-33.08 = -34.25 + 1.17)

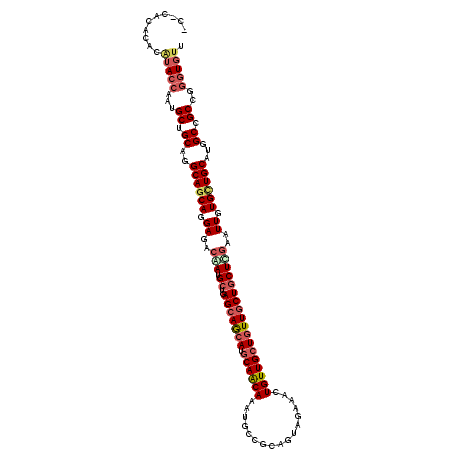
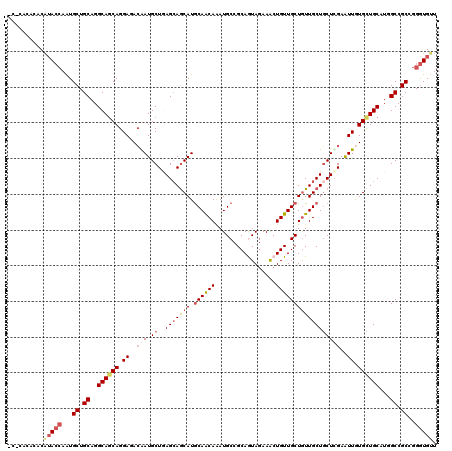
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:34:01 2006