
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,243,766 – 16,243,896 |
| Length | 130 |
| Max. P | 0.985018 |

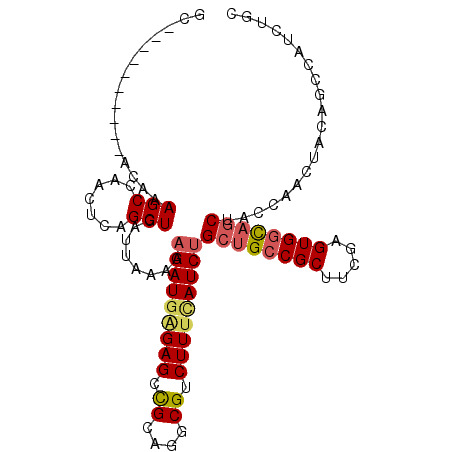
| Location | 16,243,766 – 16,243,862 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -24.90 |
| Energy contribution | -25.35 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16243766 96 + 20766785 GC----------ACAAAGCGAACUCAGCUGAUUAAAAAGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACUGCCAUCUGC ((----------(...(((.......)))........(((((((((.((....)).)))))))))((((((((.....))))))))..........)))....... ( -32.60) >DroSec_CAF1 23931 96 + 1 GC----------ACAAAGCCAACUCAGCUGAUUAAAAAGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACAGCUAUAUGC ((----------.....))......(((((.......(((((((((.((....)).)))))))))((((((((.....)))))))).........)))))...... ( -36.20) >DroSim_CAF1 24178 96 + 1 GC----------ACAAAGCCAACUCAGCUGAUUAAAAAGAUGGGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACAGCUAUAUGC ((----------.....))......(((((.......(((((..((.((....)).))..)))))((((((((.....)))))))).........)))))...... ( -35.40) >DroEre_CAF1 28982 104 + 1 GCACAUUGAGCCACUAAGCCAACUCAGCUGAUUAAAAAGAUGAGAGCCGCAGGCGUCUUUUAUCCGCUGCCGCUUCGAGUGGCAGCUACCAA--ACAGCCAUCUGC (((..(((.((......)))))....((((........((((((((.((....)).)))))))).((((((((.....))))))))......--.))))....))) ( -33.80) >DroYak_CAF1 36817 103 + 1 GCACAUUGAGCCACAAAGCCAACUCAGCUGAUUAAAAAGAUGGGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUAGCA---ACUGCCAUCUGC (((..(((.((......)))))....((((.......(((((..((.((....)).))..)))))((((((((.....)))))))))))).---..)))....... ( -35.90) >DroAna_CAF1 1341 86 + 1 CC----------ACAAAGCCAACUCAGCUGAUUAAAAAGAUCUGAGCUGGCGGCGGCUUUCAUCUGCUGACGCUCCGAGUGGUUCCUGCCAACU----------GG ((----------(....((((.(((((((........))..))))).))))(((((...((((..((....)).....))))...)))))...)----------)) ( -25.00) >consensus GC__________ACAAAGCCAACUCAGCUGAUUAAAAAGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCAACUACAGCCAUCUGC ................(((.......)))........(((((((((.((....)).)))))))))((((((((.....)))))))).................... (-24.90 = -25.35 + 0.45)

| Location | 16,243,793 – 16,243,896 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 75.72 |
| Mean single sequence MFE | -30.43 |
| Consensus MFE | -23.35 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16243793 103 + 20766785 AGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACUGCCAUCUGCUAAGUACACUCGAAAAAAAAAUA-CACAUAAA-AC--A (((((((((.((....)).)))))))))((((((((.....))))))))...(((.((((((.....))..))))...)))...........-........-..--. ( -31.60) >DroSec_CAF1 23958 101 + 1 AGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACAGCUAUAUGCUUAGUACACUCGAAAAAAAAACA-CACA--UA-GA--A (((((((((.((....)).)))))))))((((((((.....)))))))).....((((.(((.....)))))))..................-....--..-..--. ( -33.80) >DroSim_CAF1 24205 100 + 1 AGAUGGGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCGACUACAGCUAUAUGCUUAGUACACUCGAAAA-AACACA-CACA--AU-GA--A (((((..((.((....)).))..)))))((((((((.....)))))))).....((((.(((.....)))))))...........-......-....--..-..--. ( -33.00) >DroEre_CAF1 29019 98 + 1 AGAUGAGAGCCGCAGGCGUCUUUUAUCCGCUGCCGCUUCGAGUGGCAGCUACCAA--ACAGCCAUCUGCUUAGUACACUCGCGAA-A-AAUACCUCA--UC-AA--A .((((((....((((..(.(((((....((((((((.....))))))))....))--).)).)..))))...((......))...-.-.....))))--))-..--. ( -29.70) >DroYak_CAF1 36854 99 + 1 AGAUGGGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUAGCA---ACUGCCAUCUGCUUAGUACACUCGCAAAAA-AAUACUUUA--AGUUA--C (((((..((.((....)).))..)))))((((((((.....))))))))..((.---...))....(((..((....)).)))....-.........--.....--. ( -29.70) >DroAna_CAF1 1368 94 + 1 AGAUCUGAGCUGGCGGCGGCUUUCAUCUGCUGACGCUCCGAGUGGUUCCUGCCAACU----------GG-UACUACACUGUGAAAAAAAUCA-UAUGCCAA-GCUGC ((((..((((((....))))))..))))(((...((.....(((((.((........----------))-.)))))..(((((......)))-)).))..)-))... ( -24.80) >consensus AGAUGAGAGCCGCAGGCGUCUUUCAUCUGCUGCCGCUUCGAGUGGCAGCUACCAACUACAGCCAUCUGCUUAGUACACUCGAAAAAAAAACA_CACA__AA_GA__A (((((((((.((....)).)))))))))((((((((.....)))))))).......................................................... (-23.35 = -23.80 + 0.45)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:51 2006