| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,241,928 – 16,242,021 |
| Length | 93 |
| Max. P | 0.544582 |

| Location | 16,241,928 – 16,242,021 |
|---|---|
| Length | 93 |
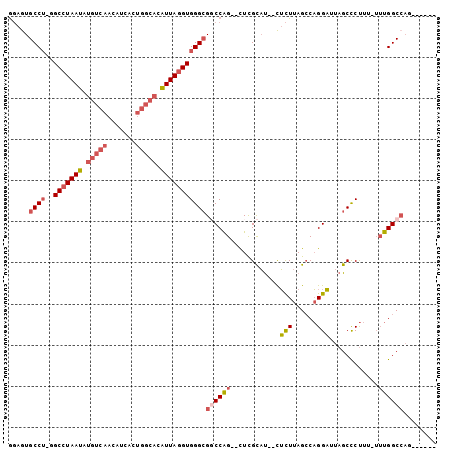
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
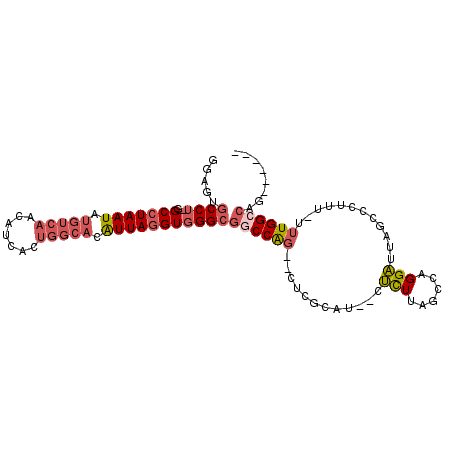
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -20.49 |
| Energy contribution | -21.80 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544582 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16241928 93 + 20766785 GGAGUGCCU-GGCCUAAUAUGUCAACAUCAGUGGCACAUUAGGUGGGCGGCCAG--UUUGCAU--CUCUUAGGCAGGAUUAGCCCUUUUUUUGGCCAG------ .....((((-(.((((((.(((((.......))))).)))))))))))((((((--..(((..--.......)))((......)).....))))))..------ ( -30.70) >DroPse_CAF1 35180 89 + 1 ACGAUGCCAGGGCCUAAUAUGUCAACAUG--------GUUAGGUGGGCGUCCGG--CUCGUCCCCUGUUUAGCCCGGCUUAGCUCC-----UGGCCAGGCCAUC ..((((.((((((((((((((....))).--------)))))))(((((.....--..)))))))))....(((.((((.......-----.)))).))))))) ( -33.60) >DroSec_CAF1 22117 92 + 1 GGAGUGCCU-GGCCUAAUAUGUCAACAUCACUGGCACAUUAGGUGGGAGUCCAG--CUCGCAU--CUCUUAGCCAGGAUUAGCCCUUU-UUUGGCCAG------ .......((-((((............(((.(((((.....(((((.(((.....--))).)))--))....)))))))).........-...))))))------ ( -31.25) >DroSim_CAF1 22388 92 + 1 GGAGUGCCU-GGCCUAAUAUGUCAACAUCACUGGCACAUUAGGUGGGCGGCCAG--CUCGCAU--CUCUUAGCCAGGAUUAGCCCUUU-UUUGGCCAG------ ((.((.(((-((((((((.(((((.......))))).)))))((((((.....)--)))))..--......))))))....))))...-.........------ ( -32.60) >DroEre_CAF1 27150 92 + 1 GGAGUGCCU-GGCCUAAUAUGUCAACUUCAAUGGCACAUUACGUGGGCGGCCAG--CUCGCAU--CUCUUAGCCAGGAUUAGCCCUUU-UUUGGCCAG------ ((.((.(((-(((.((((.(((((.......))))).)))).((((((.....)--)))))..--......))))))....))))...-.........------ ( -29.10) >DroYak_CAF1 34968 91 + 1 GGAGUGCCU-GGCCUAAUAUGUCAACAUCACUGGCACAUUAGGUGGGCGGCCAGCGCUCGCAU--CUCUUAGCCAGGAUUAGCCCUUU-UUUGGC--------- ((.((.(((-((((((((.(((((.......))))).)))))(((((((.....)))))))..--......))))))....))))...-......--------- ( -34.20) >consensus GGAGUGCCU_GGCCUAAUAUGUCAACAUCACUGGCACAUUAGGUGGGCGGCCAG__CUCGCAU__CUCUUAGCCAGGAUUAGCCCUUU_UUUGGCCAG______ .....((((..(((((((.(((((.......))))).)))))))))))((((((............(((......)))............))))))........ (-20.49 = -21.80 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:50 2006