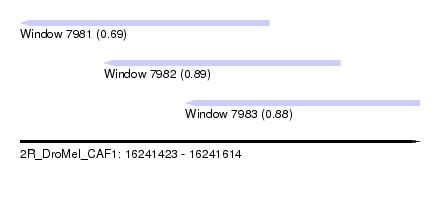
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,241,423 – 16,241,614 |
| Length | 191 |
| Max. P | 0.894046 |

| Location | 16,241,423 – 16,241,542 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.48 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16241423 119 - 20766785 UACCAAUUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUUAUUUACUAGGCUGCUUGCUGCAAUUUUUUACAC-AUAAACAUUUAAAAUGCCUAAGGAUUAUCGAAAAAUUCGCACGCAUU ....((((((((((((((.....))))))))(((((((........))))))).....))))))........-............(((((....(((((.......)))))....))))) ( -23.70) >DroSec_CAF1 21617 116 - 1 UACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUAUUUACUACGCUGCUUGCUGCAAUU---UACAC-AUAAACAUUUAAACUGCCUAAGGAUUAUCGAAAAAUUCGCUCGCAUU ....((((((((((((((.....)))))))))))))).............(((..((.(((.((---((...-.........)))).)))....(((((.......)))))))..))).. ( -23.80) >DroSim_CAF1 21888 116 - 1 UAAAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUAUUUACUACGCUGCUUGCUGCAAUU---UACAC-AUAAACAUUUAAACUGCCUAAGGAUUAUCGAAAAAUUCGCUCGCAUU ....((((((((((((((.....)))))))))))))).............(((..((.(((.((---((...-.........)))).)))....(((((.......)))))))..))).. ( -23.80) >DroEre_CAF1 26660 113 - 1 UACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUAAUUUACUUAGCUGCUUGCUGCAAUU---AGCGCAAUAAACAUUUAAAAUGCCUAAGGAUUAUCGAAAAAUACGCACG---- ......(.(((..(((((((((..(((((..((((((..........))))))((((.((....---.)))))).....)))))..)))))))))...))).).............---- ( -27.70) >DroYak_CAF1 34487 97 - 1 UACAAACUGUAUAUUUAAGCAUGCUAAAUAUGCGGCAUAAUUUACUAGGUCGCUUGCUGCAAUU---UACAC---AAACAUUUCAAAUGCCUAAGGAUUA-------------ACG---- ......(((((((((((.(....))))))))))))..((((((..((((..((.....))....---.....---...(((.....))))))).))))))-------------...---- ( -15.10) >consensus UACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUUAUUUACUACGCUGCUUGCUGCAAUU___UACAC_AUAAACAUUUAAAAUGCCUAAGGAUUAUCGAAAAAUUCGCACGCAUU .............(((((((((((.......(((((............))))).....))).............(((....)))...))))))))......................... (-14.80 = -14.48 + -0.32)

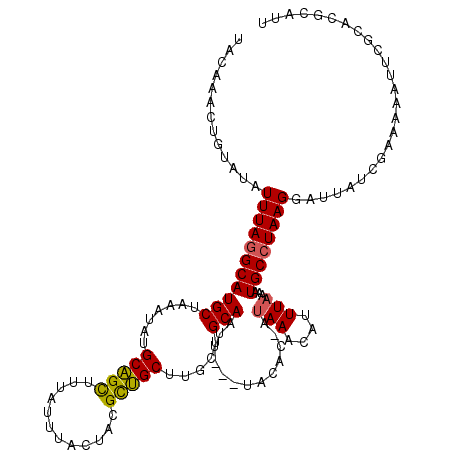
| Location | 16,241,463 – 16,241,576 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -16.46 |
| Energy contribution | -16.58 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16241463 113 - 20766785 AUUUUUUAUCUAAUUGCGCCGGUU------UUUGUUUAAUUACCAAUUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUUAUUUACUAGGCUGCUUGCUGCAAUUUUUUACAC-AUAAACA ...........((((((((.(((.------.((....))..)))......((((((((.....))))))))(((((((........)))))))..)).))))))........-....... ( -23.20) >DroSec_CAF1 21657 109 - 1 AUUUUUU-UCUAAUUGCGCCGGUU------UUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUAUUUACUACGCUGCUUGCUGCAAUU---UACAC-AUAAACA .......-...((((((((.(((.------..(((.........((((((((((((((.....)))))))))))))).........)))..))).)).))))))---.....-....... ( -25.77) >DroSim_CAF1 21928 109 - 1 AUUUUUU-UCUAAUUGCGCCGGUU------UUUGUUUAAUUAAAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUAUUUACUACGCUGCUUGCUGCAAUU---UACAC-AUAAACA .......-...((((((((.(((.------..(((.........((((((((((((((.....)))))))))))))).........)))..))).)).))))))---.....-....... ( -25.77) >DroEre_CAF1 26696 110 - 1 AUUUUUU-UGUAUUCGCCACGGUU------UUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUAAUUUACUUAGCUGCUUGCUGCAAUU---AGCGCAAUAAACA ..(((.(-(((....(((((((((------(.((......)).))))))).......)))..(((((...((((((...................)))))).))---))))))).))).. ( -23.42) >DroYak_CAF1 34510 113 - 1 AUUUUUU-UCGAAUUGCAAAGGUUUUAGUUUUUAUUUAAUUACAAACUGUAUAUUUAAGCAUGCUAAAUAUGCGGCAUAAUUUACUAGGUCGCUUGCUGCAAUU---UACAC---AAACA .......-..((((((((.((((((((((........(((((....(((((((((((.(....))))))))))))..))))).))))))..))))..)))))))---)....---..... ( -22.10) >consensus AUUUUUU_UCUAAUUGCGCCGGUU______UUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUUAUUUACUACGCUGCUUGCUGCAAUU___UACAC_AUAAACA ...........((((((.............................((((((((((((.....)))))))))))).............((.....)).))))))................ (-16.46 = -16.58 + 0.12)

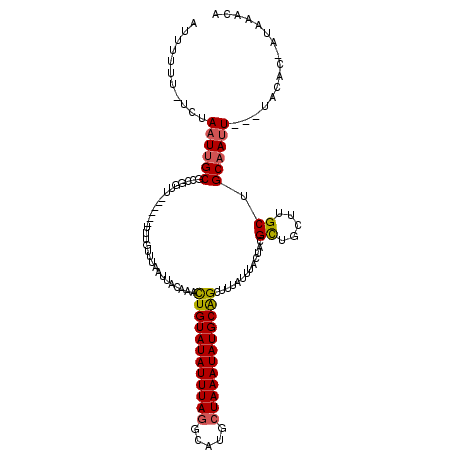

| Location | 16,241,502 – 16,241,614 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.04 |
| Mean single sequence MFE | -24.05 |
| Consensus MFE | -18.04 |
| Energy contribution | -17.91 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16241502 112 - 20766785 AA--AGCUAAUUAAUUUGUGUUUCUGUUUUUUCGAUAAAUAUUUUUUAUCUAAUUGCGCCGGUUUUUGUUUAAUUACCAAUUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUUA ((--((((.........((((....(((.....((((((.....)))))).))).)))).(((..((....))..)))....((((((((((.....)))))))))))))))). ( -21.30) >DroSec_CAF1 21693 113 - 1 AAUUCGCUAAUUAAUUCGUGUUUCUGCGCUUUCGAUAAAUAUUUUUU-UCUAAUUGCGCCGGUUUUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUA .....(.(((((((..((.(..((.((((.((.((.(((....))).-)).))..)))).))..).)).)))))))).((((((((((((((.....))))))))))))))... ( -26.00) >DroSim_CAF1 21964 111 - 1 AA--AGCUAAUUAAUUCGUGUUUCUGCGCUUUCGAUAAAUAUUUUUU-UCUAAUUGCGCCGGUUUUUGUUUAAUUAAAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGUUAUA ..--...(((((((..((.(..((.((((.((.((.(((....))).-)).))..)))).))..).)).)))))))..((((((((((((((.....))))))))))))))... ( -26.00) >DroEre_CAF1 26733 111 - 1 AA--AGCUAAUUAAUUCGAGUUGUUGGGUUUUUGAUAAAUAUUUUUU-UGUAUUCGCCACGGUUUUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUUAA .(--((((((((((..((((..(..((((.....(((((......))-)))....))).)..).)))).))))))).....(((((((((((.....))))))))))))))).. ( -22.90) >consensus AA__AGCUAAUUAAUUCGUGUUUCUGCGCUUUCGAUAAAUAUUUUUU_UCUAAUUGCGCCGGUUUUUGUUUAAUUACAAACUGUAUAUUUAGGCAUGCUAAAUAUGCAGCUAUA .......(((((((.........((((((....((.(((.....))).)).....))).))).......)))))))..((((((((((((((.....))))))))))))))... (-18.04 = -17.91 + -0.13)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:49 2006