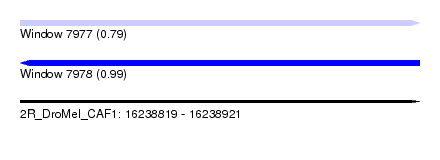
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,238,819 – 16,238,921 |
| Length | 102 |
| Max. P | 0.989989 |

| Location | 16,238,819 – 16,238,921 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -18.68 |
| Consensus MFE | -11.44 |
| Energy contribution | -11.67 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
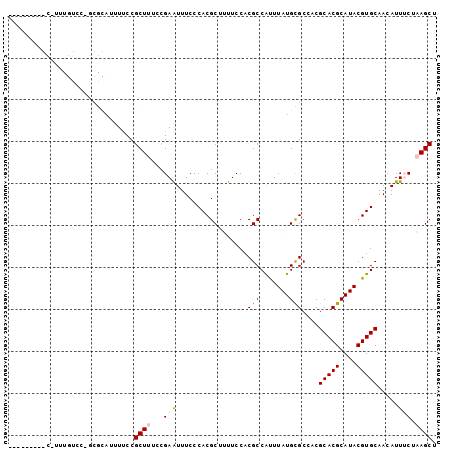
>2R_DroMel_CAF1 16238819 102 + 20766785 UUUCCUCAGCUUUUGUGCCGCGCAUUUUCCGCUUUUCGAAUUUCCCACGCUUUUCCACGCCAUUUAUGCGCCACGCACGCAUACGUGCAACAUUUCUAAGCU .......(((((..(((..((((((.....((.....(((............)))...)).....)))))))))(((((....))))).........))))) ( -22.00) >DroPse_CAF1 32126 79 + 1 -----------------------GGCUCCCGCUCAGUGAGUUUCCCACGCAAUUCCACGCCGCUUAUGUGCCACGCACGCAUACGUGCAACAUUGCUAAGCU -----------------------((((........(((.......)))(((((.....(.(((....))).)..(((((....)))))...)))))..)))) ( -18.00) >DroSec_CAF1 19144 102 + 1 UUUCCUCAGCUUUUGUCCUGCGCAUUUUCCGCUUUCCGAAUUUCCCACGCUUUUCCACGCCAUUUAUGCGCCACGCACGCAUACGUGCAACAUUUCUAAGCU .......(((((.(((...((((((.....((.....(((............)))...)).....))))))...(((((....))))).))).....))))) ( -20.00) >DroEre_CAF1 24208 93 + 1 ---------UCUUUCCCCAGCGCAUUUUCCGCUUUCCGAAUUUCCCACGCUUUUCCACGCCAUUUAUGCGCCACGCACGCAUACGUGCAACAUUUCUAAGCU ---------..........((((((.....((.....(((............)))...)).....))))))...(((((....))))).............. ( -16.30) >DroYak_CAF1 31935 102 + 1 UUUCCUCAGCCUUUGUCCUGCGCAUUUUCCGCUUUUCGAAUUUCCCACGCUUUUCCACGCCAUUUAUGCGCCACGCACGCAUACGUGCAACAUUUCUAAGCU .......(((.........((((((.....((.....(((............)))...)).....))))))...(((((....)))))...........))) ( -17.80) >DroPer_CAF1 32090 79 + 1 -----------------------GGCUCCCGCUCAGUGAGUUUCCCACGCAAUUCCACGCCGCUUAUGUGCCACGCACGCAUACGUGCAACAUUGCUAAGCU -----------------------((((........(((.......)))(((((.....(.(((....))).)..(((((....)))))...)))))..)))) ( -18.00) >consensus _________C_UUUGUCC_GCGCAUUUUCCGCUUUCCGAAUUUCCCACGCUUUUCCACGCCAUUUAUGCGCCACGCACGCAUACGUGCAACAUUUCUAAGCU ..............................((((...(((.................(((.......)))....(((((....))))).....))).)))). (-11.44 = -11.67 + 0.22)

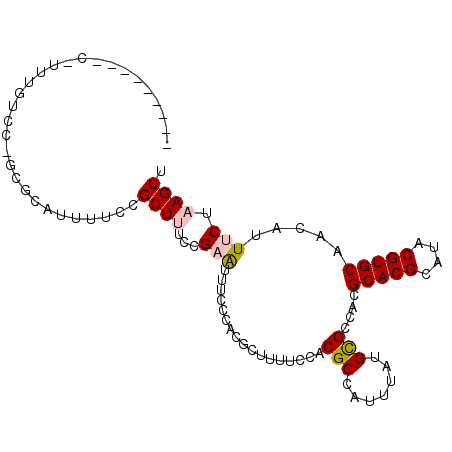
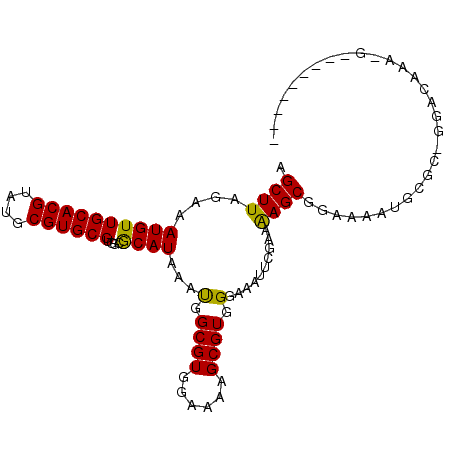
| Location | 16,238,819 – 16,238,921 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -18.83 |
| Energy contribution | -18.17 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.989989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16238819 102 - 20766785 AGCUUAGAAAUGUUGCACGUAUGCGUGCGUGGCGCAUAAAUGGCGUGGAAAAGCGUGGGAAAUUCGAAAAGCGGAAAAUGCGCGGCACAAAAGCUGAGGAAA ..(((((...(((.(((((....)))))((.((((((...(.((((......)))).)....((((.....))))..)))))).)))))....))))).... ( -32.80) >DroPse_CAF1 32126 79 - 1 AGCUUAGCAAUGUUGCACGUAUGCGUGCGUGGCACAUAAGCGGCGUGGAAUUGCGUGGGAAACUCACUGAGCGGGAGCC----------------------- .((((((((((...(((((....)))))((.((......)).)).....)))))((((....).)))))))).......----------------------- ( -29.30) >DroSec_CAF1 19144 102 - 1 AGCUUAGAAAUGUUGCACGUAUGCGUGCGUGGCGCAUAAAUGGCGUGGAAAAGCGUGGGAAAUUCGGAAAGCGGAAAAUGCGCAGGACAAAAGCUGAGGAAA ..(((((...(((((((((....)))))...((((((...(.((((......)))).)....((((.....))))..))))))..))))....))))).... ( -31.10) >DroEre_CAF1 24208 93 - 1 AGCUUAGAAAUGUUGCACGUAUGCGUGCGUGGCGCAUAAAUGGCGUGGAAAAGCGUGGGAAAUUCGGAAAGCGGAAAAUGCGCUGGGGAAAGA--------- ..(((.........(((((....)))))..(((((((...(.((((......)))).)....((((.....))))..))))))).....))).--------- ( -24.80) >DroYak_CAF1 31935 102 - 1 AGCUUAGAAAUGUUGCACGUAUGCGUGCGUGGCGCAUAAAUGGCGUGGAAAAGCGUGGGAAAUUCGAAAAGCGGAAAAUGCGCAGGACAAAGGCUGAGGAAA ..(((((...(((((((((....)))))...((((((...(.((((......)))).)....((((.....))))..))))))..))))....))))).... ( -31.00) >DroPer_CAF1 32090 79 - 1 AGCUUAGCAAUGUUGCACGUAUGCGUGCGUGGCACAUAAGCGGCGUGGAAUUGCGUGGGAAACUCACUGAGCGGGAGCC----------------------- .((((((((((...(((((....)))))((.((......)).)).....)))))((((....).)))))))).......----------------------- ( -29.30) >consensus AGCUUAGAAAUGUUGCACGUAUGCGUGCGUGGCGCAUAAAUGGCGUGGAAAAGCGUGGGAAAUUCGAAAAGCGGAAAAUGCGC_GGACAAA_G_________ .((((....((((((((((....))))))....))))...(.((((......)))).)..........)))).............................. (-18.83 = -18.17 + -0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:44 2006