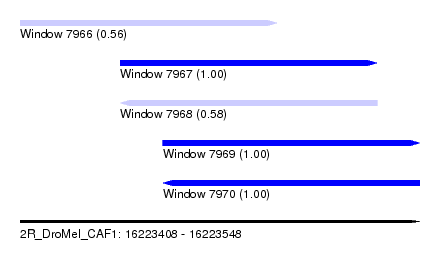
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,223,408 – 16,223,548 |
| Length | 140 |
| Max. P | 0.997937 |

| Location | 16,223,408 – 16,223,498 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 84.88 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -16.61 |
| Energy contribution | -17.80 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16223408 90 + 20766785 GCGAAAGUUGUCAGGCAGUUAAUAAGCCGACUCUUUUGUGGUAGUUGGUUGUAGCUCAGCAGCUCAGUAGCUCAGUAGCUCAGUAGCUGU ((....))......(.(((((...((((((((((.....)).)))))))).)))))).((((((....((((....))))....)))))) ( -26.20) >DroSec_CAF1 4440 82 + 1 GCGAAAGUUGUCAGGCAGUUAAUAAGCUGACUCUUUUGUGGUAGUUGGCUGUAGCUC--------AGUUGCUCAGUAGCUUAGUAGCUGU ((....))......(((((((.((((((((((((.....)).)))).((((.(((..--------....))))))))))))).))))))) ( -26.50) >DroSim_CAF1 4488 82 + 1 GCGAAAGUUGUCAGGCAGUUAAUAAGCUGACUCUUUUGUGGUAGUUGGCUGUAGCUC--------AGUUGCUCAGUAGCUCAGUAGCUGU ((....))........(((((...(((..(((((.....)).)))..))).)))))(--------(((((((.((...)).)))))))). ( -23.70) >DroYak_CAF1 15677 73 + 1 GCGAAAGUUGUCAGGCAGUUAAUAAGCUGACUCUUUUGUGGUAGUUGGUAGUUGG-----------------UAGUAGCUCAGUAGCUGC (((((((..(((((............))))).))))))).(((((((.((((((.-----------------...))))).).))))))) ( -20.10) >consensus GCGAAAGUUGUCAGGCAGUUAAUAAGCUGACUCUUUUGUGGUAGUUGGCUGUAGCUC________AGUUGCUCAGUAGCUCAGUAGCUGU ((....))......(((((((...((((((((((.....)).))))))))((((((.........))))))............))))))) (-16.61 = -17.80 + 1.19)

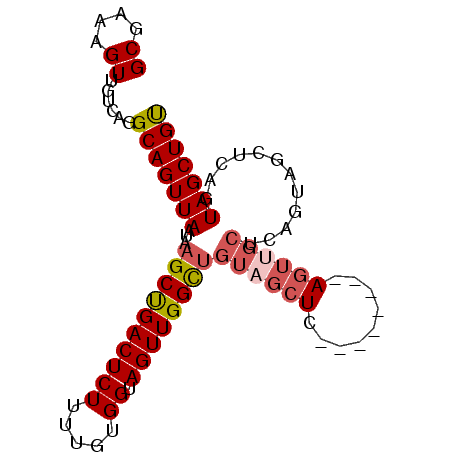
| Location | 16,223,443 – 16,223,533 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -26.30 |
| Energy contribution | -26.17 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997259 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16223443 90 + 20766785 UUGUGGUAGUUGGUUGUAGCUCAGCAGCUCAGUAGCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAU .(((...((((..((((((..((((((((....((((....))))....))))))))..(((....)))...))))))..))))..))). ( -32.30) >DroSec_CAF1 4475 82 + 1 UUGUGGUAGUUGGCUGUAGCUC--------AGUUGCUCAGUAGCUUAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAU .(((...(((..((((((((.(--------(((((((.((...)).))))))))..)).(((....)))....))))))..)))..))). ( -35.20) >DroSim_CAF1 4523 82 + 1 UUGUGGUAGUUGGCUGUAGCUC--------AGUUGCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAU .(((...(((..((((((((.(--------(((((((.((...)).))))))))..)).(((....)))....))))))..)))..))). ( -35.20) >DroYak_CAF1 15712 73 + 1 UUGUGGUAGUUGGUAGUUGG-----------------UAGUAGCUCAGUAGCUGCUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAU .(((...(((..(..((..(-----------------((((((((....))))))))).(((....))).....))..)..)))..))). ( -27.30) >consensus UUGUGGUAGUUGGCUGUAGCUC________AGUUGCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAU .(((...(((..(((((((..................((((((((....))))))))..(((....)))...)))))))..)))..))). (-26.30 = -26.17 + -0.12)

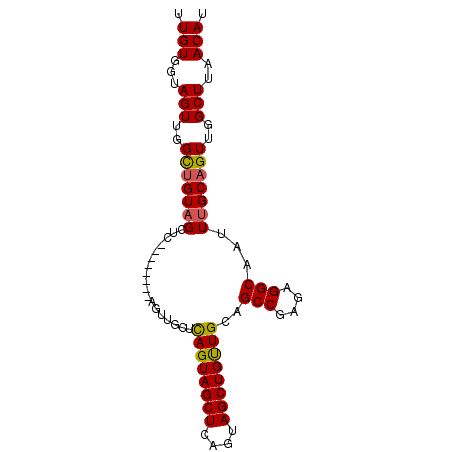
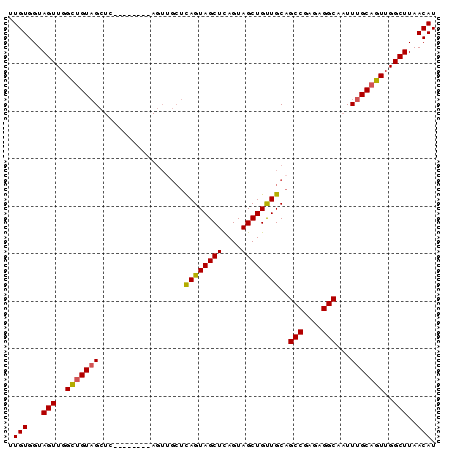
| Location | 16,223,443 – 16,223,533 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 85.47 |
| Mean single sequence MFE | -16.88 |
| Consensus MFE | -11.46 |
| Energy contribution | -11.27 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580539 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16223443 90 - 20766785 AUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGCUACUGAGCUGCUGAGCUACAACCAACUACCACAA .(((..(((((..((((....(((....))))))).(((((....((((....))))....))))).)).)))))).............. ( -22.10) >DroSec_CAF1 4475 82 - 1 AUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUAAGCUACUGAGCAACU--------GAGCUACAGCCAACUACCACAA ..(((.((((....((.....)).....))))..)))((((....)))).((((((....--------..))).)))............. ( -16.50) >DroSim_CAF1 4523 82 - 1 AUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGCAACU--------GAGCUACAGCCAACUACCACAA ..(((.((((....((.....)).....))))..)))((((....)))).((((((....--------..))).)))............. ( -16.60) >DroYak_CAF1 15712 73 - 1 AUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAGCAGCUACUGAGCUACUA-----------------CCAACUACCAACUACCACAA ..(((.......(((((....(((....)))))))).((((....))))....-----------------..)))............... ( -12.30) >consensus AUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGCAACU________GAGCUACAACCAACUACCACAA ..(((.((((....((.....)).....))))..)))((((....))))......................................... (-11.46 = -11.27 + -0.19)

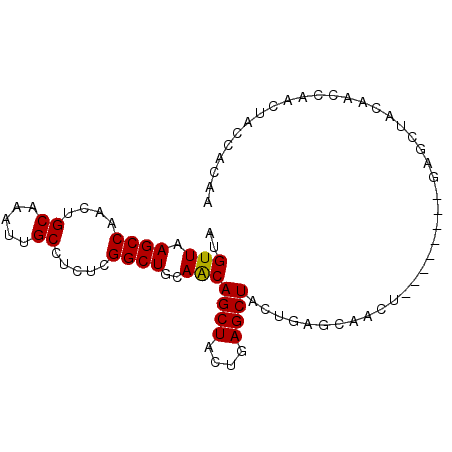
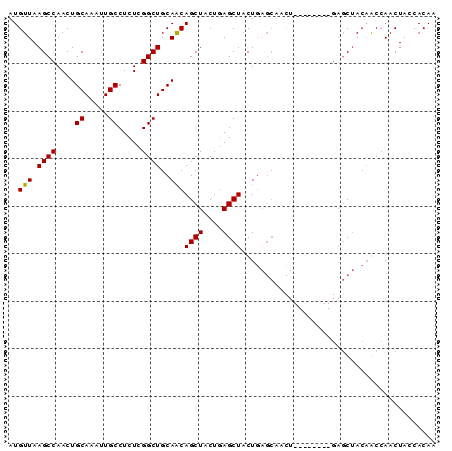
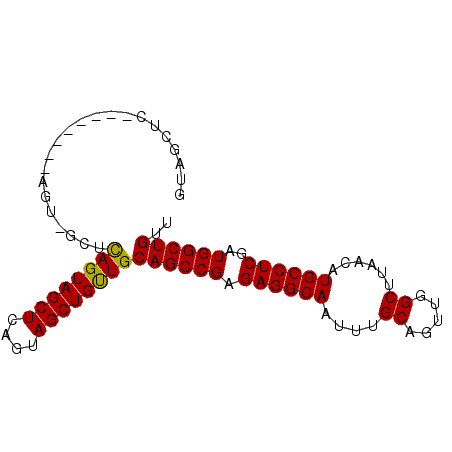
| Location | 16,223,458 – 16,223,548 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -30.06 |
| Consensus MFE | -26.18 |
| Energy contribution | -25.78 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.97 |
| SVM RNA-class probability | 0.997937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16223458 90 + 20766785 GUAGCUCAGCAGCUCAGUAGCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU ......((((((((....((((....))))....))))))))((((((.((((((....((.....))......))))))..)))))).. ( -34.00) >DroSec_CAF1 4490 82 + 1 GUAGCUC--------AGUUGCUCAGUAGCUUAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU ......(--------(((((((.((...)).))))))))..(((((((.((((((....((.....))......))))))..))))))). ( -30.10) >DroSim_CAF1 4538 82 + 1 GUAGCUC--------AGUUGCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU ......(--------(((((((.((...)).))))))))..(((((((.((((((....((.....))......))))))..))))))). ( -30.10) >DroEre_CAF1 9237 71 + 1 --UGG-----------------UAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU --..(-----------------((((((((....)))))))))(((((.((((((....((.....))......))))))..)))))... ( -25.40) >DroYak_CAF1 15727 73 + 1 GUUGG-----------------UAGUAGCUCAGUAGCUGCUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU ....(-----------------((((((((....)))))))))(((((.((((((....((.....))......))))))..)))))... ( -30.70) >consensus GUAGCUC________AGU_GCUCAGUAGCUCAGUAGCUGUUGCAGCCGAGAGGCAAUUUGCAGUUGGCUUAACAUGCCUCGAUGGCUGUU ......................((((((((....))))))))((((((.((((((....((.....))......))))))..)))))).. (-26.18 = -25.78 + -0.40)

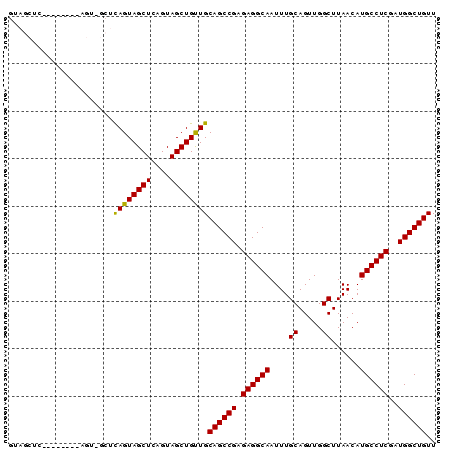
| Location | 16,223,458 – 16,223,548 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 85.53 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -20.98 |
| Energy contribution | -20.98 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16223458 90 - 20766785 AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGCUACUGAGCUGCUGAGCUAC ..(((((...((((((......((.....))....))))))..)))))((.(((((....((((....))))....))))).))...... ( -31.00) >DroSec_CAF1 4490 82 - 1 AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUAAGCUACUGAGCAACU--------GAGCUAC ..(((((...((((((......((.....))....))))))..)))))....((((.....(((....)))....--------.)))).. ( -24.40) >DroSim_CAF1 4538 82 - 1 AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGCAACU--------GAGCUAC ..(((((...((((((......((.....))....))))))..)))))....((((.....(((....)))....--------.)))).. ( -24.40) >DroEre_CAF1 9237 71 - 1 AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUA-----------------CCA-- ..(((((...((((((......((.....))....))))))..)))))....((((....))))....-----------------...-- ( -21.00) >DroYak_CAF1 15727 73 - 1 AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAGCAGCUACUGAGCUACUA-----------------CCAAC ..(((((...((((((......((.....))....))))))..))))).((.((((....)))).)).-----------------..... ( -22.00) >consensus AACAGCCAUCGAGGCAUGUUAAGCCAACUGCAAAUUGCCUCUCGGCUGCAACAGCUACUGAGCUACUGAGC_ACU________GAGCUAC ..(((((...((((((......((.....))....))))))..)))))....((((....)))).......................... (-20.98 = -20.98 + 0.00)

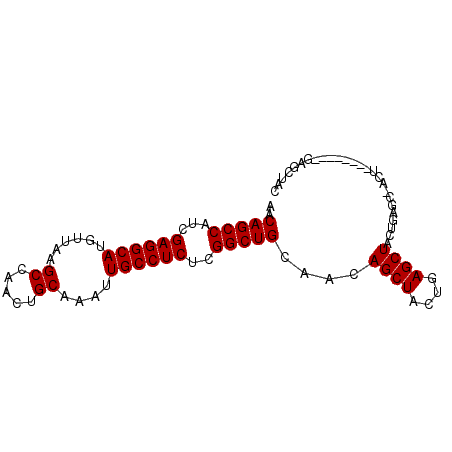
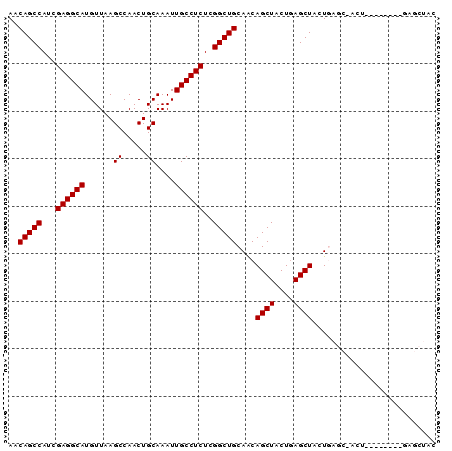
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:37 2006