| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,220,646 – 16,220,741 |
| Length | 95 |
| Max. P | 0.645013 |

| Location | 16,220,646 – 16,220,741 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -17.32 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645013 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16220646 95 + 20766785 UAUGCAACAUCGGCACUGCACUGCAACUGCGUGCAGCUGUGUGUGUG------------------CUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGCAGAAACA ..(((.((((..(((((((((.((....)))))))).)))..))))(------------------((((((((((....))))))((((.....))))))))).)))...... ( -33.60) >DroPse_CAF1 13926 81 + 1 UAUGCAACAGCGG--CGGUAAUGCAAU-G------UUUGUG--UG---------------------UGUGUGCGUGUGAGUGCAUUGGCAAGUGGCCAAGAGCGGCAACAGCU ...((....))((--((.(((((((..-.------((((..--((---------------------(....)))..))))))))))).)..((.(((......))).)).))) ( -21.50) >DroSec_CAF1 1697 91 + 1 UAUGCAACAUCGG--CUGCACUGCAACUGCGUGCAGCUGUG--UGUG------------------CUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGCAGAAACA ..(((.(((((((--((((((.((....))))))))))).)--)))(------------------((((((((((....))))))((((.....))))))))).)))...... ( -37.60) >DroSim_CAF1 1701 91 + 1 UAUGCAACAUCGG--CUGCACUGCAACUGCGUGCAGCUGUG--UGUG------------------CUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGCAGAAACA ..(((.(((((((--((((((.((....))))))))))).)--)))(------------------((((((((((....))))))((((.....))))))))).)))...... ( -37.60) >DroEre_CAF1 6436 81 + 1 -AUGCAAC-----------ACUGCAACUGCGUGCAGCUGUG--UGUG------------------CUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGCAAAAACA -.(((..(-----------((((....((((..((((....--...)------------------)))..))))..........(((((.....)))))))))))))...... ( -26.20) >DroYak_CAF1 12747 108 + 1 -AUGCAACAUCGG--CGGCACUGCAACUGCGUGCAGCUGUG--UGUGCUGUGAGUGCAGCGUGUGCUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGUAAAAACA -..((((((.(((--(.((((.((....)))))).))))))--).)))(((.((((((.((.((((.....)))).))..)))))).))).((.((((....))))....)). ( -39.10) >consensus UAUGCAACAUCGG__CUGCACUGCAACUGCGUGCAGCUGUG__UGUG__________________CUGUGUGUAUUCGAGUGCAUUGGCAAGUGGCCAACAGUGGCAAAAACA ..(((............((((.((....)))))).(((((.............................((((((....))))))((((.....))))))))).)))...... (-17.32 = -18.27 + 0.95)


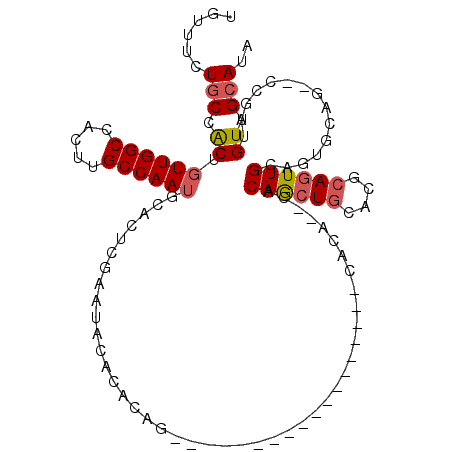
| Location | 16,220,646 – 16,220,741 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 79.24 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.53 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16220646 95 - 20766785 UGUUUCUGCCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAG------------------CACACACACAGCUGCACGCAGUUGCAGUGCAGUGCCGAUGUUGCAUA (((((((((....((((((.....)))))))))...))).)))....(------------------((.((((...(((((((((....)).)))))))...).))))))... ( -26.90) >DroPse_CAF1 13926 81 - 1 AGCUGUUGCCGCUCUUGGCCACUUGCCAAUGCACUCACACGCACACA---------------------CA--CACAAA------C-AUUGCAUUACCG--CCGCUGUUGCAUA .((....((.((..(((((.....)))))(((........)))....---------------------..--......------.-...........)--).))....))... ( -14.50) >DroSec_CAF1 1697 91 - 1 UGUUUCUGCCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAG------------------CACA--CACAGCUGCACGCAGUUGCAGUGCAG--CCGAUGUUGCAUA (((((((((....((((((.....)))))))))...))).)))....(------------------((.(--(((.(((((((((....)).))))))--).).))))))... ( -29.00) >DroSim_CAF1 1701 91 - 1 UGUUUCUGCCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAG------------------CACA--CACAGCUGCACGCAGUUGCAGUGCAG--CCGAUGUUGCAUA (((((((((....((((((.....)))))))))...))).)))....(------------------((.(--(((.(((((((((....)).))))))--).).))))))... ( -29.00) >DroEre_CAF1 6436 81 - 1 UGUUUUUGCCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAG------------------CACA--CACAGCUGCACGCAGUUGCAGU-----------GUUGCAU- (((.((((.....((((((.....)))))).....)))).)))..(((------------------(((.--..((((((....))))))..))-----------))))...- ( -23.80) >DroYak_CAF1 12747 108 - 1 UGUUUUUACCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAGCACACGCUGCACUCACAGCACA--CACAGCUGCACGCAGUUGCAGUGCCG--CCGAUGUUGCAU- .............((((((.....))))))(((((((........((((....))))........((((.--..((((((....))))))..))))..--.))).).)))..- ( -29.20) >consensus UGUUUCUGCCACUGUUGGCCACUUGCCAAUGCACUCGAAUACACACAG__________________CACA__CACAGCUGCACGCAGUUGCAGUGCAG__CCGAUGUUGCAUA ......(((.((.((((((.....))))))............................................((((((....))))))...............)).))).. (-14.10 = -14.53 + 0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:32 2006