| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,219,655 – 16,219,759 |
| Length | 104 |
| Max. P | 0.566448 |

| Location | 16,219,655 – 16,219,759 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -20.99 |
| Energy contribution | -21.52 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
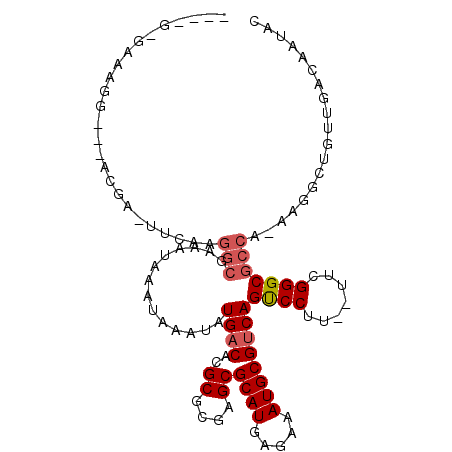
>2R_DroMel_CAF1 16219655 104 - 20766785 ----GGGAAAGGGACGCGA-UUCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCCUUAUUCGGGCGCCA-AAGGCUGUUGACAAUAC ----(((.(((((((((..-......))..............((((..((....))((((.....))))))))))))))).))).((((((.-..))).)))........ ( -31.70) >DroSec_CAF1 698 98 - 1 ----G-GGAAGG---ACGA-UUCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU--UUGGGGCGCCA-AAGGCUGUUGACAAUAC ----.-((((((---((..-(((.....)))...........((((..((....))((((.....))))))))))))))--))..((((((.-..))).)))........ ( -27.20) >DroSim_CAF1 719 98 - 1 ----G-GAAAGG---ACGA-UUCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU--UUGGGGCGCCA-AAGGCUGUUGACAAUAC ----.-((((((---((..-(((.....)))...........((((..((....))((((.....))))))))))))))--))..((((((.-..))).)))........ ( -27.60) >DroEre_CAF1 5463 99 - 1 ----A-AAAGGG---ACGA-UUCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCCUCCU-CGGGCGCCA-AAGGCUGUUGACAAAAG ----.-...(((---((..-(((.....)))...........((((..((....))((((.....)))))))))))))((..-(((.(....-..).)))..))...... ( -28.00) >DroYak_CAF1 11821 100 - 1 ----A-AAAGGG---ACAA-UUCCCUGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUUCUUUUGGGCGCCA-AAGGCUGUUGACAAUAC ----.-..((((---(...-.)))))................((((..((....))((((.....))))))))((((.......))))(((.-..)))............ ( -27.60) >DroMoj_CAF1 38574 100 - 1 GGGGC-CUG--A---ACAUUCCCAAGACGAAAUAAAUAAGUAUGACACGCGCGAGCGCAUGAGAAAUGCGGCAGCCCUU----UGGGCCACACAAACGUGUUGACAAUUC ..(((-(..--(---(.....((.........................((....))((((.....))))))......))----..))))((((....))))......... ( -24.20) >consensus ____G_GAAAGG___ACGA_UUCAAGGCGAAAUAAAUAAAUAUGACACGCGCGAGCGCAUGAGAAAUGCGUCAGUCCUU__UUCGGGCGCCA_AAGGCUGUUGACAAUAC .........................(((..............((((..((....))((((.....))))))))((((.......)))))))................... (-20.99 = -21.52 + 0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:28 2006