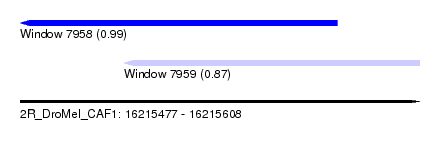
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,215,477 – 16,215,608 |
| Length | 131 |
| Max. P | 0.993981 |

| Location | 16,215,477 – 16,215,581 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 72.28 |
| Mean single sequence MFE | -24.09 |
| Consensus MFE | -6.21 |
| Energy contribution | -7.33 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.26 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
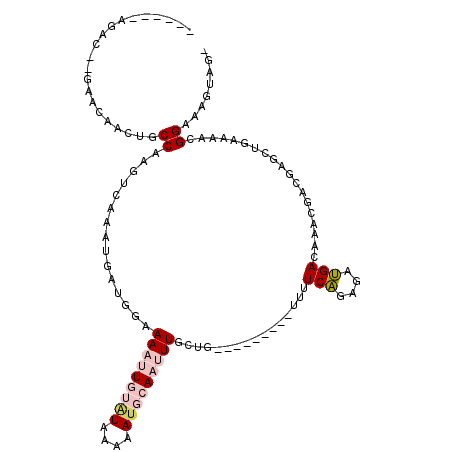
>2R_DroMel_CAF1 16215477 104 - 20766785 GGAAAAUUGUAUAAAAAUGCAAUUUACUG---------UUUUCAGAGAUGACAAACGACGAGCUGAAAACGGAAAGUAG---AAAAUUGCUAGCCGCUGAAAGCUAAAAAGUGCAA ((.(((((((((....))))))))).))(---------(((((((.(.((.....)).)...))))))))((..(((((---....)))))..)).......((........)).. ( -21.80) >DroPse_CAF1 8218 111 - 1 GGAAAAUUAUGUGAAAAUUCAUGUUGAUGGCUUGUUUCUUCUCUGCCUGGACAAACAAA-AGC-AAAGAGGGAGAGAAGGGAAAACAUUCUGU---UUGUAAGCUGAAUAUGGCAA ..................(((((((..((((((((..(((((((.(((...........-...-....))).)))))))..)((((.....))---)).))))))))))))))... ( -25.97) >DroEre_CAF1 1338 101 - 1 GGAAAAUUGUAUAAAAAUGCAAUUUACUG---------UUUUCAGAGAUGACAAACGGCGAGCUGAAAACGGAAACUAG---AAAAUUGCUAG---CUGAAAGCUAAAAAGUGCAA ......((((((......(((((((.(((---------(((((((.(.((.....)).)...))))))))(....).))---.)))))))(((---(.....))))....)))))) ( -24.00) >DroYak_CAF1 7708 101 - 1 GGAAAAUUGUAUAAAAAUGCAAUUUGCUG---------UUUUCAGUGAUGACAAACGGCGGGCUGAAAACGGAAAGUAG---AAAAUUGCUAG---CUGAAAGCUAAAAAGUGCAA ......((((((......(((((((.(((---------((((((((..((........)).))))))))))).......---.)))))))(((---(.....))))....)))))) ( -25.80) >DroPer_CAF1 8115 111 - 1 GGAAAAUUAUUUGAAAAUUCAUGUUGAUGACUUGUUUCUUCUCUGCCUGGACAAACAAA-AGC-AAAGAGGGAGAGAAGGGAAAACAUUCUGU---UUGUAAGCUGAAUAUGGCAA ..................(((((((.(.(.(((((..(((((((.(((...........-...-....))).)))))))..)((((.....))---)).)))))).)))))))... ( -22.87) >consensus GGAAAAUUGUAUAAAAAUGCAAUUUGCUG_________UUUUCAGAGAUGACAAACGACGAGCUGAAAACGGAAAGUAG___AAAAUUGCUAG___CUGAAAGCUAAAAAGUGCAA ..................(((((((.............((((((((...............))))))))..............)))))))............((........)).. ( -6.21 = -7.33 + 1.12)

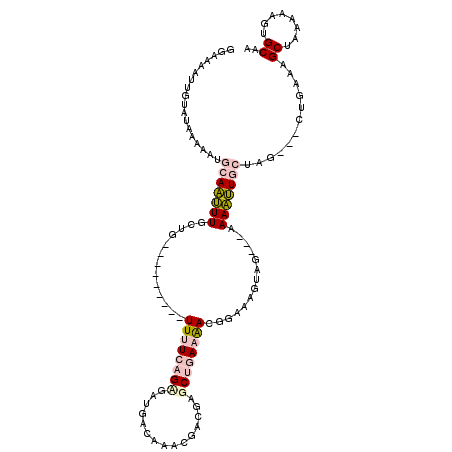
| Location | 16,215,511 – 16,215,608 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 72.21 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -4.26 |
| Energy contribution | -5.46 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.874454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16215511 97 - 20766785 ------AGAC--GAACAACUGCCAAGUCAAAUGAUGGAAAAUUGUAUAAAAAUGCAAUUUACUG---------UUUUCAGAGAUGACAAACGACGAGCUGAAAACGGAAAGUAG- ------....--......((((...(((....)))...(((((((((....))))))))).(((---------(((((((.(.((.....)).)...))))))))))...))))- ( -20.40) >DroPse_CAF1 8251 113 - 1 GACGACGGACGAGAACAAUUUCCAUGGAAAAUUAUGGAAAAUUAUGUGAAAAUUCAUGUUGAUGGCUUGUUUCUUCUCUGCCUGGACAAACAAA-AGC-AAAGAGGGAGAGAAGG ......(((((((.....(((((((((....)))))))))(((((((((....))))).))))..)))))))(((((((.(((...........-...-....))).))))))). ( -27.57) >DroEre_CAF1 1369 97 - 1 ------AGAC--GAACAACUGCCAAGUCAAAUGAUGGAAAAUUGUAUAAAAAUGCAAUUUACUG---------UUUUCAGAGAUGACAAACGGCGAGCUGAAAACGGAAACUAG- ------.(((--.............)))......(((.(((((((((....))))))))).(((---------(((((((.(.((.....)).)...))))))))))...))).- ( -20.42) >DroYak_CAF1 7739 97 - 1 ------AGAC--GAACAACUGCCAAGUCAAAUGAUGGAAAAUUGUAUAAAAAUGCAAUUUGCUG---------UUUUCAGUGAUGACAAACGGCGGGCUGAAAACGGAAAGUAG- ------...(--(.......(((..(((....)))...(((((((((....)))))))))((((---------((((((....))).))))))).)))......))........- ( -22.62) >DroPer_CAF1 8148 113 - 1 GACGACGGACGAGAACAAUUGCCAUGGAAAAUUAUGGAAAAUUAUUUGAAAAUUCAUGUUGAUGACUUGUUUCUUCUCUGCCUGGACAAACAAA-AGC-AAAGAGGGAGAGAAGG ......(((((((........((((((....))))))................((((....)))))))))))(((((((.(((...........-...-....))).))))))). ( -23.47) >consensus ______AGAC__GAACAACUGCCAAGUCAAAUGAUGGAAAAUUGUAUAAAAAUGCAAUUUGCUG_________UUUUCAGAGAUGACAAACGACGAGCUGAAAACGGAAAGUAG_ .....................((...............(((((((((....)))))))))................(((....)))...................))........ ( -4.26 = -5.46 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:27 2006