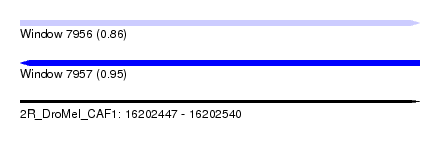
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,202,447 – 16,202,540 |
| Length | 93 |
| Max. P | 0.948847 |

| Location | 16,202,447 – 16,202,540 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -18.62 |
| Energy contribution | -20.54 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16202447 93 + 20766785 AAGGAGUGGAAAAGCUGUCAGAUUAAAUGCCAUAAUCCAUGGGAUUAUGGCCAAGCUGGCAAAAGUUGUUAAUGCCCGGUGACAGAGCUCAAC-- ...((((.......(((((.........((((((((((...))))))))))...(((((((...........)).)))))))))).))))...-- ( -30.00) >DroSec_CAF1 16698 93 + 1 AAGGAGUGGAAAAGCUGUCAGAUUAAAUGCCAUAAUCCAUGGGAUUAUGGCCAAGCUGGCAAAAGUUGUUAAUGCCCGGUGACAGAGCUCAAC-- ...((((.......(((((.........((((((((((...))))))))))...(((((((...........)).)))))))))).))))...-- ( -30.00) >DroSim_CAF1 16583 93 + 1 AAGGAGUGGAAAAGCUGUCAGAUUAAAUGCCAUAAUCCAUGGGAUUAUGGCCAAGCUGGCAAAAGUUGUUAAUGCCCGGUGACAGAGCUCAAC-- ...((((.......(((((.........((((((((((...))))))))))...(((((((...........)).)))))))))).))))...-- ( -30.00) >DroEre_CAF1 16830 92 + 1 AACGGGUGGAAGAGCUGUCAGAUUAAAUGCCAUAAUCCUAUGGAUUAUGGCCAAGCUGG-AAAAGUUGUUAAUACCCGGUGACAGAGCUCAAC-- ..((((((.((.((((.((((.......((((((((((...))))))))))....))))-...)))).))..))))))...............-- ( -31.30) >DroYak_CAF1 17403 93 + 1 AACGGGUGGAAAAGCUGUCAGAUUAAAUGCCAUAAUCCAUGGGAUUAUGGCCAAGCUGGUAAAAGUUGUUAAUGCCCGGUGACAGAGCUCAAC-- ..((((((....((((.((((.......((((((((((...))))))))))....))))....)))).....))))))...............-- ( -28.40) >DroPer_CAF1 19816 82 + 1 AAGC-CAGAGAGAG-----AGAAAAAAUGGAUGUAUCCAGGGGAUUUUGGCAAGGGUGACAGAAGU-------ACCUGGUGACAGGCCUCAAAAG ..((-(((((....-----........((((....)))).....)))))))...(....)......-------.((((....))))......... ( -19.03) >consensus AAGGAGUGGAAAAGCUGUCAGAUUAAAUGCCAUAAUCCAUGGGAUUAUGGCCAAGCUGGCAAAAGUUGUUAAUGCCCGGUGACAGAGCUCAAC__ ...((((.......(((((.........((((((((((...))))))))))...(((((................)))))))))).))))..... (-18.62 = -20.54 + 1.92)

| Location | 16,202,447 – 16,202,540 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -25.07 |
| Consensus MFE | -19.99 |
| Energy contribution | -21.38 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16202447 93 - 20766785 --GUUGAGCUCUGUCACCGGGCAUUAACAACUUUUGCCAGCUUGGCCAUAAUCCCAUGGAUUAUGGCAUUUAAUCUGACAGCUUUUCCACUCCUU --((.(((..((((((...((((...........))))......((((((((((...))))))))))........))))))...))).))..... ( -28.60) >DroSec_CAF1 16698 93 - 1 --GUUGAGCUCUGUCACCGGGCAUUAACAACUUUUGCCAGCUUGGCCAUAAUCCCAUGGAUUAUGGCAUUUAAUCUGACAGCUUUUCCACUCCUU --((.(((..((((((...((((...........))))......((((((((((...))))))))))........))))))...))).))..... ( -28.60) >DroSim_CAF1 16583 93 - 1 --GUUGAGCUCUGUCACCGGGCAUUAACAACUUUUGCCAGCUUGGCCAUAAUCCCAUGGAUUAUGGCAUUUAAUCUGACAGCUUUUCCACUCCUU --((.(((..((((((...((((...........))))......((((((((((...))))))))))........))))))...))).))..... ( -28.60) >DroEre_CAF1 16830 92 - 1 --GUUGAGCUCUGUCACCGGGUAUUAACAACUUUU-CCAGCUUGGCCAUAAUCCAUAGGAUUAUGGCAUUUAAUCUGACAGCUCUUCCACCCGUU --...(((((..((((.(((((.............-...)))))((((((((((...))))))))))........)))))))))........... ( -26.59) >DroYak_CAF1 17403 93 - 1 --GUUGAGCUCUGUCACCGGGCAUUAACAACUUUUACCAGCUUGGCCAUAAUCCCAUGGAUUAUGGCAUUUAAUCUGACAGCUUUUCCACCCGUU --(..(((((..((((.(((((.................)))))((((((((((...))))))))))........)))))))))..)........ ( -26.63) >DroPer_CAF1 19816 82 - 1 CUUUUGAGGCCUGUCACCAGGU-------ACUUCUGUCACCCUUGCCAAAAUCCCCUGGAUACAUCCAUUUUUUCU-----CUCUCUCUG-GCUU ......(((((((.((...(((-------((....)).)))..)).))........((((....))))........-----........)-)))) ( -11.40) >consensus __GUUGAGCUCUGUCACCGGGCAUUAACAACUUUUGCCAGCUUGGCCAUAAUCCCAUGGAUUAUGGCAUUUAAUCUGACAGCUUUUCCACUCCUU .....(((((..((((.(((((.................)))))((((((((((...))))))))))........)))))))))........... (-19.99 = -21.38 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:25 2006