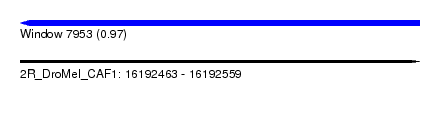
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,192,463 – 16,192,559 |
| Length | 96 |
| Max. P | 0.967847 |

| Location | 16,192,463 – 16,192,559 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 70.82 |
| Mean single sequence MFE | -23.25 |
| Consensus MFE | -9.25 |
| Energy contribution | -9.08 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967847 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16192463 96 - 20766785 ----GGCCAUUCU--GAAAUUAUAUCGUAUCCAAUCUGCAUAACAAAUG-CAUAACUAGAUGCCCGUUUUCAAGGCCAAAGCCAAUGGGCAAUCA-AAUACGUA ----.........--..........(((((......(((((.....)))-))......((((((((((.....(((....)))))))))).))).-.))))).. ( -21.80) >DroSec_CAF1 6784 80 - 1 ----GGCCAUUCU--GAAAUCAUGUCGGAUUCAAUCUGCAUAACAAAUG-CAGAACCAGAUGUACGUUUUCAAGGCCAAAGCCAAU-----------------A ----((((..(((--((.......))))).....(((((((.....)))-))))...................)))).........-----------------. ( -20.70) >DroSim_CAF1 6626 80 - 1 ----GGCCAUUCU--GAAAUCAUGUCGGAUUCAAUCUGCAUAACAAAUG-CAGAACCAGGUGCACGUUUUCAAGGCCAAAGCCAAU-----------------A ----((((..(((--((.......))))).....(((((((.....)))-))))...................)))).........-----------------. ( -20.70) >DroEre_CAF1 6711 92 - 1 UGUUGGCCAUUCU---AAAACAUGUCGGGUUCAAUCUGCAUAACAAAUG-CAGAACUCAAAACCCGUUUUCAAGGCCAAGGCC-------AAUCA-AAUACGUA .(((((((.....---.........((((((...(((((((.....)))-))))......)))))).............))))-------)))..-........ ( -25.73) >DroYak_CAF1 6925 97 - 1 UGUUGGCCAUUCU--GAAAACAUGUCGCUGCCAAUCUGCAUAGCAAAUG-UGGAACUCAAAGCCCGUUUUCAAGGCCAAAGCCA---GGCAAUCA-AAUACGUA ((...(((....(--((((((.....(((.....((..(((.....)))-..))......)))..))))))).(((....))).---)))...))-........ ( -23.30) >DroAna_CAF1 6652 101 - 1 UGUCGGGUGUUUUGGGAAACUAUGCCAGAUCCAAUCUGCAUAACAAAUGGCGGAAUGGGAGGCUCGUUUUCAAGGCCGAAGACAA---AAAAUGGGUAUAUGUU ((((....((((((..((.....(((...((((.(((((..........))))).)))).)))...))..))))))....)))).---................ ( -27.30) >consensus ____GGCCAUUCU__GAAAUCAUGUCGGAUCCAAUCUGCAUAACAAAUG_CAGAACUAGAUGCCCGUUUUCAAGGCCAAAGCCAA_____AAUCA_AAUACGUA ....((((..........................(((((((.....))).))))...........(....)..))))........................... ( -9.25 = -9.08 + -0.16)

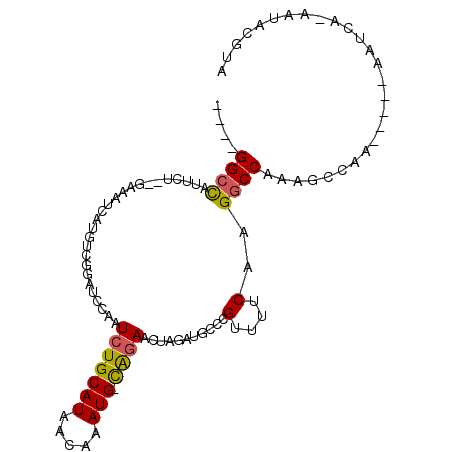
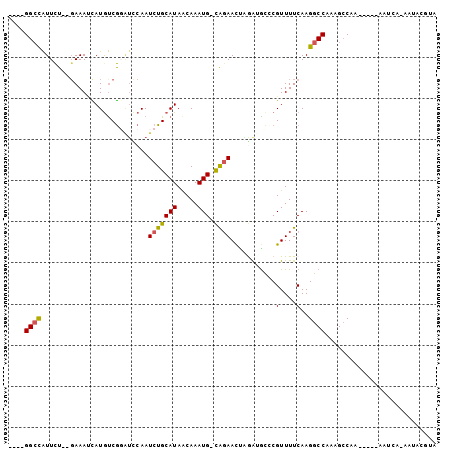
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:21 2006