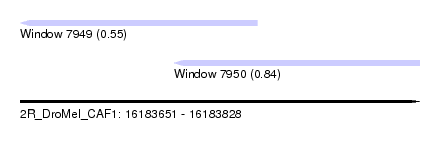
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,183,651 – 16,183,828 |
| Length | 177 |
| Max. P | 0.843707 |

| Location | 16,183,651 – 16,183,756 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.86 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -19.19 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545205 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
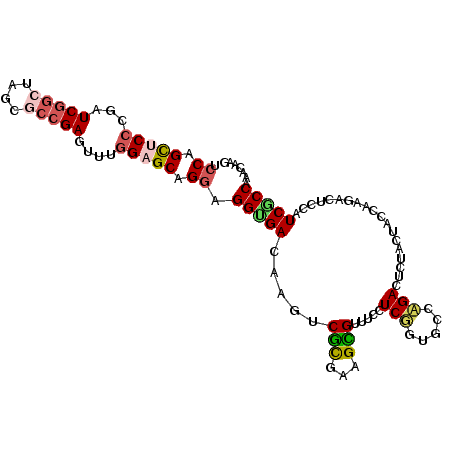
>2R_DroMel_CAF1 16183651 105 - 20766785 UCCAGCUCCCCAUCGGCUAGCACCGAGUUUGGAGCAGGAGGUGACAAGUCGCGCAGCGUUUCCUCGUUGCCCGACUCUACUACCAAGACUCCCUCGCCAAACAAG (((.(((((...((((......))))....))))).)))(((((...((((.((((((......)))))).))))(((.......))).....)))))....... ( -38.80) >DroVir_CAF1 36750 105 - 1 AGCGGUUCACGUUCCGCUAGCGAUGAAUUCGGCGCCGGCGGGGACAAAUCACAAAGUGUCUCUUCUGUGCCAGAUUCUACCACCAAAACACCGUCGCCCAUCAAA (((((........))))).(((((((((..(((((.((.(((((((..........))))))).)))))))..))))...............)))))........ ( -32.26) >DroSec_CAF1 27541 105 - 1 GCCAGCUCCCCAUCGGAGAGUACCGAGUUUGGAGCAGGAGGUGACAAGUCGUGCAGCGUUUCCUCGGUGCCCGACUCCACUACCAAGACACCCUCGCCAAACAAG ((..(((((...((((......))))....))))).((.((((...(((((.(((.((......)).))).))))).)))).))...........))........ ( -36.00) >DroEre_CAF1 28138 105 - 1 UCCAGCUCCCGUUCGGCUAGUGCCGAGUUUGGAGCAGGAGGCGACAAGUCGCGUAGCGUUUCCUCGGUGCCAGACUCUACAACCAAGACUCCAUCGCCAAACAAG (((.(((((..((((((....))))))...))))).)))(((((..((((..((((.((((..........)))).))))......))))...)))))....... ( -38.10) >DroYak_CAF1 28014 105 - 1 UCCAGUUCCCGAUCGGCUAGUGCCGAGUUUGGAGCAGGAGGUGACAAGUCGCGUAGUGUGUCCUCUGUGCCAGACUCCACUACCAAGACUCCAUCACCAAACAAG (((.(((((...(((((....)))))....))))).)))(((((..((((..((((((.(...((((...))))..)))))))...))))...)))))....... ( -41.00) >DroAna_CAF1 47453 105 - 1 GCCAGUUCCCGUUCGGCUAGCGCCGAAUUUGGUGCCGGAGGUGAUAAAUCGCAAAGCGUAUCCUCUGUACCAGAUUCAACCACUAAGUCGCCGUCACCGAAUAAG ....((((.....(((((((.(..(((((((((((.(((((........(((...)))...))))))))))))))))...).)))....)))).....))))... ( -31.20) >consensus UCCAGCUCCCGAUCGGCUAGCGCCGAGUUUGGAGCAGGAGGUGACAAGUCGCGAAGCGUUUCCUCGGUGCCAGACUCUACUACCAAGACUCCAUCGCCAAACAAG .((.(((((...(((((....)))))....))))).)).(((((.....(((...))).....(((.....)))...................)))))....... (-19.19 = -19.50 + 0.31)

| Location | 16,183,719 – 16,183,828 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.33 |
| Mean single sequence MFE | -36.07 |
| Consensus MFE | -23.57 |
| Energy contribution | -23.55 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
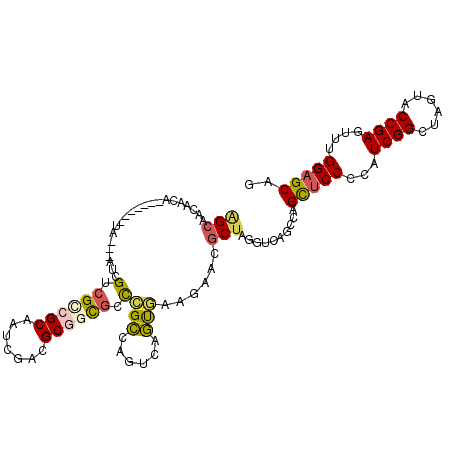
>2R_DroMel_CAF1 16183719 109 - 20766785 AGCAACAAUAAUAACAAUU---AUCGUCGUCGCAAUCGACGCGGCGGCCGCCAAUCGGUGAAGGACGCUAGGCCAUCCAGCUCCCCAUCGGCUAGCACCGAGUUUGGAGCAG (((................---...((((((((.......))))))))((((....))))......))).((....)).(((((...((((......))))....))))).. ( -35.20) >DroSec_CAF1 27609 112 - 1 GGCAACAACAAUAACAAUAACAAUCGUCGCCGCAAUCGACGCGGCGGCCGCCAUUCAGUGAAGAACGCUAGGUCAGCCAGCUCCCCAUCGGAGAGUACCGAGUUUGGAGCAG (((......................((((((((.......)))))))).(((....((((.....)))).)))..))).(((((...((((......))))....))))).. ( -39.10) >DroSim_CAF1 27813 112 - 1 GGCAACAACAAUAACAAUAACAAUCGUCGCCGCAAUCGACGCGGCGGCCGCCAUUCGGUGAAGAACGCUAGGUCAGCCAGCUCCCCAUCGGAGAGUACCGAGUUUGGAGCAG (((......................((((((((.......))))))))((((....))))...............))).(((((...((((......))))....))))).. ( -40.40) >DroEre_CAF1 28206 100 - 1 AGCACCAGCA------------AUCGUCGCCGCAAUCGACGCAAUGCCGGCCAGACAGUCAAGAACGCUAUGCCAUCCAGCUCCCGUUCGGCUAGUGCCGAGUUUGGAGCAG .((.((.(((------------..(((((.......)))))...))).))((((((......(((((....((......))...)))))(((....)))..)))))).)).. ( -32.90) >DroYak_CAF1 28082 100 - 1 AGCAACAACA------------AUCGUCGCCGCAAUCGUCGCGGCGCCGGCCAGUCGGCCAAGAACGCCAUGUCAUCCAGUUCCCGAUCGGCUAGUGCCGAGUUUGGAGCAG .((.......------------...(.((((((.......)))))).)((((....))))......))...........(((((...(((((....)))))....))))).. ( -38.10) >DroAna_CAF1 47521 92 - 1 -----------------UAACAACCGCCGCCGCAAUCGUCGCACUACUGGUCCGCCAACAAAGAAUG---GAGCGGCCAGUUCCCGUUCGGCUAGCGCCGAAUUUGGUGCCG -----------------........((....)).......((((((((((.((((...((.....))---..)))))))))....(((((((....)))))))..))))).. ( -30.70) >consensus AGCAACAACA_______UA___AUCGUCGCCGCAAUCGACGCGGCGCCCGCCAGUCAGUGAAGAACGCUAGGUCAGCCAGCUCCCCAUCGGCUAGUACCGAGUUUGGAGCAG (((......................(.((((((.......)))))).)(((......)))......)))..........(((((...((((......))))....))))).. (-23.57 = -23.55 + -0.02)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:19 2006