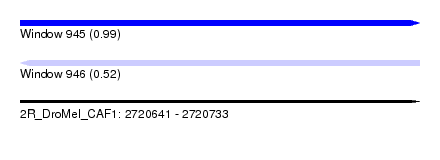
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 2,720,641 – 2,720,733 |
| Length | 92 |
| Max. P | 0.988626 |

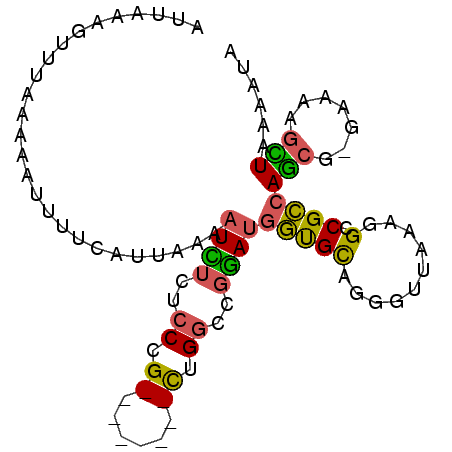
| Location | 2,720,641 – 2,720,733 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -2.96 |
| Energy contribution | -3.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.13 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2720641 92 + 20766785 UAUUUUAGCUUUUC-CGCUGGCGGCCUUUAACCCUGCACCAUCCGGCCAGCGGGAAAGCGGGAGAAAUUUUAAUGAAAAUUUU-CAACUUUAAU .......(((((((-(((((((((..................)).))))))))))))))..(((((.((((...)))).))))-)......... ( -29.37) >DroSec_CAF1 12309 94 + 1 UAUUCUAGCUUUUCCCGCUGGCGGCCUUUAACCCUGCACCAUCCGGCCAGCGGGAAAGCGGGAGAGAUUUUAAUGAAAAUUUUCCAACUUUAAU .......(((((.(((((((((((..................)).))))))))))))))((((((..(((....)))..))))))......... ( -32.27) >DroEre_CAF1 3770 81 + 1 UUUUUUAACUUUUC-CGCUGGCGGCCUUUAACCCUGCACCGUC----CAG--------CUGGAGAGAUUUUAAUGAAAAAUGUUAAACUUUAAU ...(((((((((((-(((((((((..............))).)----)))--------).))))))(((((.....)))))))))))....... ( -19.94) >DroYak_CAF1 12581 84 + 1 UACUUUAGCUUUUC-CGCUGGCGGCCUUUAACCCGGCACCAUCCGGCCAG--------CGGGAGAGAUUUUAAUUAAAUGU-UUAAACUUUAAU ........((((((-(((((((((.......))(((......))))))))--------))))))))......((((((.((-....)))))))) ( -27.30) >DroAna_CAF1 12008 83 + 1 UGUUUUAACUUUUC-UCUUGCCGACCUUUAAACCCACGAGAUACCGCCGA--------CGGGAAAUAUUUUAAUUAA--UUCGAAAACUUUAAU .(((((......((-((.((..............)).))))..(((....--------)))................--....)))))...... ( -7.14) >consensus UAUUUUAGCUUUUC_CGCUGGCGGCCUUUAACCCUGCACCAUCCGGCCAG________CGGGAGAGAUUUUAAUGAAAAUUUUUAAACUUUAAU ........((((((.(((((((.......................)))))........))))))))............................ ( -2.96 = -3.60 + 0.64)

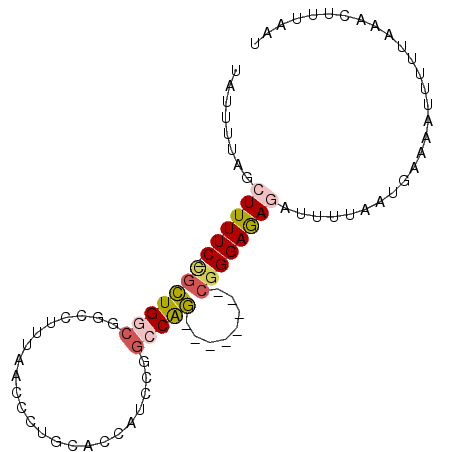
| Location | 2,720,641 – 2,720,733 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.60 |
| Mean single sequence MFE | -22.10 |
| Consensus MFE | -4.23 |
| Energy contribution | -3.74 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.19 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.520958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 2720641 92 - 20766785 AUUAAAGUUG-AAAAUUUUCAUUAAAAUUUCUCCCGCUUUCCCGCUGGCCGGAUGGUGCAGGGUUAAAGGCCGCCAGCG-GAAAAGCUAAAAUA .........(-(((.(((.....))).))))....(((((.((((((((.((....((......))....)))))))))-).)))))....... ( -25.30) >DroSec_CAF1 12309 94 - 1 AUUAAAGUUGGAAAAUUUUCAUUAAAAUCUCUCCCGCUUUCCCGCUGGCCGGAUGGUGCAGGGUUAAAGGCCGCCAGCGGGAAAAGCUAGAAUA ...((((((....))))))............((..((((((((((((((.((....((......))....))))))))))).)))))..))... ( -29.50) >DroEre_CAF1 3770 81 - 1 AUUAAAGUUUAACAUUUUUCAUUAAAAUCUCUCCAG--------CUG----GACGGUGCAGGGUUAAAGGCCGCCAGCG-GAAAAGUUAAAAAA .......(((((((((((.....)))))...(((.(--------(((----(.((((............))))))))))-))...))))))... ( -19.90) >DroYak_CAF1 12581 84 - 1 AUUAAAGUUUAA-ACAUUUAAUUAAAAUCUCUCCCG--------CUGGCCGGAUGGUGCCGGGUUAAAGGCCGCCAGCG-GAAAAGCUAAAGUA .....(((((..-..((((.....)))).....(((--------((((((((......)))(((.....))))))))))-)..)))))...... ( -23.80) >DroAna_CAF1 12008 83 - 1 AUUAAAGUUUUCGAA--UUAAUUAAAAUAUUUCCCG--------UCGGCGGUAUCUCGUGGGUUUAAAGGUCGGCAAGA-GAAAAGUUAAAACA ....((.(((((...--................(((--------....)))..(((.((.(..(....)..).)).)))-))))).))...... ( -12.00) >consensus AUUAAAGUUUAAAAAUUUUCAUUAAAAUCUCUCCCG________CUGGCCGGAUGGUGCAGGGUUAAAGGCCGCCAGCG_GAAAAGCUAAAAUA ..........................((((..((.((......)).))..))))(((((..........).))))(((.......)))...... ( -4.23 = -3.74 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:12 2006