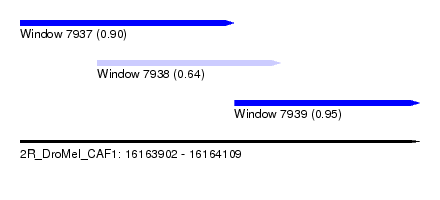
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,163,902 – 16,164,109 |
| Length | 207 |
| Max. P | 0.954718 |

| Location | 16,163,902 – 16,164,013 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -29.39 |
| Consensus MFE | -28.11 |
| Energy contribution | -28.31 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
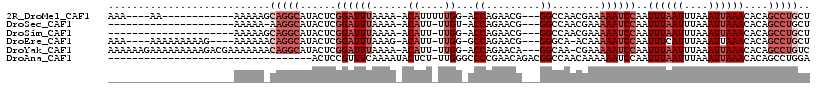
>2R_DroMel_CAF1 16163902 111 + 20766785 GGAGCGAACAGACCAGAACCCAAACAUUGUCCAUAAAUUUCGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A--C (((((((((.(((...............)))....((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....)))))..----.--. ( -25.76) >DroSec_CAF1 7982 110 + 1 GGAGCGAACGGACCAGAACCCAAACAAUGUCCAUAAAUUUCGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A--- (((((((((((((...............))))...((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....)))))..----.--- ( -30.06) >DroSim_CAF1 8439 110 + 1 GGAGCGAACGGACCAGAACCCAAACAAUGUCCAUAAAUUUCGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A--- (((((((((((((...............))))...((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....)))))..----.--- ( -30.06) >DroEre_CAF1 7935 113 + 1 GGAGCGAACGGACCAGAGCCCAAACAAUGUCCAUAAAUUUUGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----AAAC (((((((((((((...............))))...((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....)))))..----.... ( -30.86) >DroYak_CAF1 7957 113 + 1 GGAGCGAACGGACCAGAACCCAAACAAUGUCCAUAAAUUUUGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACAAACC---- (((((((((((((...............))))...((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....)))))......---- ( -30.86) >DroAna_CAF1 8275 108 + 1 GGAGCGGACGGACCAGAACCGAAACACUGUCCAUAAAUUUCGAGCUCAAAUUGCUUUUAGUUGUUCGGAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUAC--------- .(.((.(((((.((.(..(((((..((((............((((.......)))).))))..)))))..)))...))))).))).......(((((.....))))).--------- ( -28.72) >consensus GGAGCGAACGGACCAGAACCCAAACAAUGUCCAUAAAUUUCGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA____A___ (((((((((((((...............))))...((((..((((.......))))..))))))))))...(((((....)))))...))).(((((.....))))).......... (-28.11 = -28.31 + 0.20)

| Location | 16,163,942 – 16,164,037 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -23.48 |
| Consensus MFE | -16.93 |
| Energy contribution | -16.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640313 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16163942 95 + 20766785 CGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A--CAAA----AA------------AAAAAGCAGGCAUACUCGG ((((......((((((((.((((....))))(..((((((..((.(((....)))))..))))))..).----.--....----..------------.)))))))).....)))). ( -27.00) >DroSec_CAF1 8022 88 + 1 CGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A------------------------AAAAA-AAGGCAUACUCGG ((((.......(((((((.((((....))))(..((((((..((.(((....)))))..))))))..).----.------------------------....)-))))))..)))). ( -23.90) >DroSim_CAF1 8479 89 + 1 CGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----A------------------------AAAAAGCAGGCAUACUCGG ((((......((((((((.((((....))))(..((((((..((.(((....)))))..))))))..).----.------------------------.)))))))).....)))). ( -27.00) >DroEre_CAF1 7975 105 + 1 UGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA----AAACAAA----AAAAAAAAAG----AAAAAACAGGCAUACUCGG ...(((.......(((((.((((....))))(..((((((..((.(((....)))))..))))))..).----.......----.....)))))----........)))........ ( -19.76) >DroYak_CAF1 7997 113 + 1 UGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACAAACC----AAAAAAGAAAAAAAAAGACGAAAAAAACAGGCAUACUCGG .(((.......(((((...(((.(((((...(((((....))))).......(((((.....)))))......----................).))))...))))))))..))).. ( -20.70) >DroAna_CAF1 8315 74 + 1 CGAGCUCAAAUUGCUUUUAGUUGUUCGGAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUAC-------------------------------------------ACUCCG .((((.......)))).........(((((.(..((((((..((.(((....)))))..))))))..)-------------------------------------------.))))) ( -22.50) >consensus CGAGCUCAAAUUGCUUUUAGUUGUUCGCAGCGGCUGCCGUCGGCCAUAUUCAUAUGCAAAUGGCAUACA____A________________________AAAAA_CAGGCAUACUCGG ((((..............((((((.....))))))(((((..((.(((....)))))..)))))................................................)))). (-16.93 = -16.93 + 0.00)

| Location | 16,164,013 – 16,164,109 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.32 |
| Mean single sequence MFE | -17.78 |
| Consensus MFE | -6.05 |
| Energy contribution | -7.88 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
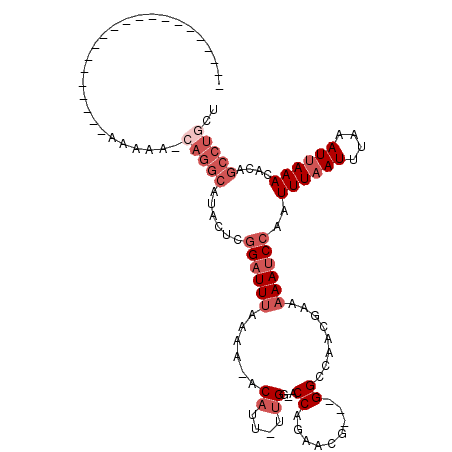
>2R_DroMel_CAF1 16164013 96 + 20766785 AAA----AA------------AAAAAGCAGGCAUACUCGGAUUUAAAA-ACAUUUUUGG-ACCAGAACG---GGCCAACGAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGCU ...----..------------....(((((((......((((((....-...(((((((-.((......---)))))).)))))))))..((((((....))))))....))))))) ( -22.81) >DroSec_CAF1 8092 89 + 1 ---------------------AAAAA-AAGGCAUACUCGGAUUUAAAA-ACAUU-UUGU-ACCAGAACG---GGCCAACGAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGCU ---------------------.....-.((((......((((((....-....(-((((-.((.....)---)....)))))))))))..((((((....))))))....))))... ( -13.70) >DroSim_CAF1 8549 90 + 1 ---------------------AAAAAGCAGGCAUACUCGGAUUUAAAA-ACAUU-UUGG-ACCAGAACG---GGCCAACGAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGCU ---------------------....(((((((......((((((....-.....-((((-.((......---))))))....))))))..((((((....))))))....))))))) ( -22.72) >DroEre_CAF1 8048 102 + 1 AAA----AAAAAAAAAG----AAAAAACAGGCAUACUCGGAUUUAAAG-ACAUU-UUGG-GCCAGAACG---GGGCA-ACAAAAAUCCAAUUUCAUUUAAAUUAAACACAGCCUGCU ...----..........----......(((((.......((((((((.-.....-((((-(((.....)---)(...-.).....))))).....)))))))).......))))).. ( -19.14) >DroYak_CAF1 8070 110 + 1 AAAAAAGAAAAAAAAAGACGAAAAAAACAGGCAUACUCGGAUUUAAAA-ACAUU-UUGG-ACCAGAACA---GGCAA-CGAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGUC ..........................((((((.......((((((((.-.....-((((-(........---(....-)......))))).....)))))))).......)))))). ( -18.58) >DroAna_CAF1 8383 82 + 1 ----------------------------------ACUCCGUUUCAAAAUACUCU-UUGGGCCCCGAACAGACGGCCAACAAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGGA ----------------------------------...................(-(..(((.(((......)))................((((((....))))))....)))..)) ( -9.70) >consensus _____________________AAAAA_CAGGCAUACUCGGAUUUAAAA_ACAUU_UUGG_ACCAGAACG___GGCCAACGAAAAAUCCAAUUUAAUUUAAAUUAAACACAGCCUGCU ...........................(((((......((((((......((....))...((.........))........))))))..((((((....))))))....))))).. ( -6.05 = -7.88 + 1.83)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:33:08 2006