
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,131,682 – 16,131,784 |
| Length | 102 |
| Max. P | 0.763416 |

| Location | 16,131,682 – 16,131,784 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 81.93 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -18.25 |
| Energy contribution | -20.03 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16131682 102 - 20766785 U-----------GACGCAAUCAUUUGCAUCUUAUUUUAAUUCCUAUGAUUGUUUCACUGCCAGGCGGAAUAACCAGUUGCUUUUUGUCUUGUUGUUUGCUGCUUGGCU---GUUGU .-----------((.((((((((.....................)))))))).))((.(((((((((.(((((.((..((.....)))).)))))...))))))))).---))... ( -28.00) >DroPse_CAF1 30465 110 - 1 UCAUCUUAAAGUGACGCAAUCAUUUGAAUAUUAUUUCGAUUCCUAUGAUUGUUUCACUCUCUGGCGGAAUAACCAGUUGCUUGCUGGCUGGCUGUUC---GUUUCGUU---GUUGU .........(((((.((((((((..((((.........))))..)))))))).)))))....(((((((..(.((((((((....))).))))).).---.)))))))---..... ( -30.60) >DroSec_CAF1 20753 109 - 1 UGAAC-CAAAGUGACGCAAUCAUUUGCAUCUUAUUUUAAUUCCUAUGAUUGUUUCACUGCCAGGCGGAAUAACCAGUUGCUUUUUGUCGU---GUUUGCAGCUUGGCU---GUUGU .....-...(((((.((((((((.....................)))))))).)))))(((((((((.....))...(((..........---....)))))))))).---..... ( -28.64) >DroEre_CAF1 21322 112 - 1 UGAAC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCCAGGCUGAAUAACCAGUUGCUGUUUGUCGUGUUGUUUGCUGCUUGGCU---GUUGU .....-...(((((.((((((((..(.(((.......))).)..)))))))).)))))(((((((.(((((((.....((.....))...)))))))...))))))).---..... ( -30.40) >DroYak_CAF1 21067 115 - 1 UGAAC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCCAGGCUGAAUAACCAGUUGCUGCUUGUCGUGUUGUUUGCUGCUUGGCUGAUGUUGU .....-...(((((.((((((((..(.(((.......))).)..)))))))).)))))(((((((.(((((((((((....)))....).)))))))...)))))))......... ( -30.70) >DroAna_CAF1 21701 104 - 1 UCACC-CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUUCUAUGAUUUUUUCAUUGGAUGGCUAAAUAACCAGUUGC-------CGUGUUGUUU-CUAUUUGGCU---GUUGU .((.(-((((..((.((((.(((..(((..((((((.....(((((((.....))).)))).....)))))).....)))-------.))))))).)-)..))))).)---).... ( -18.70) >consensus UGAAC_CAAAGUGACGCAAUCAUUUGCAUUUUAUUUUAAUUCCUAUGAUUGUUUCACUGCCAGGCGGAAUAACCAGUUGCUUUUUGUCGUGUUGUUUGCUGCUUGGCU___GUUGU .........(((((.((((((((.....................)))))))).)))))(((((((.(((((((.................)))))))...)))))))......... (-18.25 = -20.03 + 1.78)

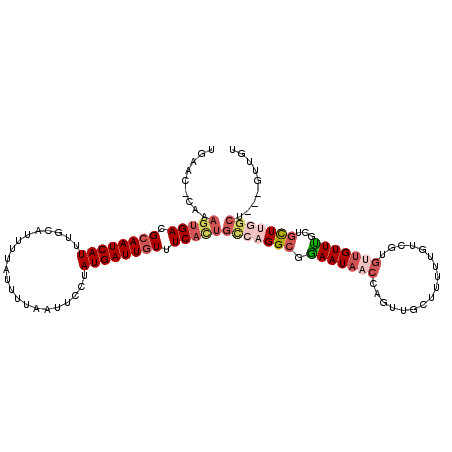
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:58 2006