
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,100,167 – 16,100,303 |
| Length | 136 |
| Max. P | 0.997123 |

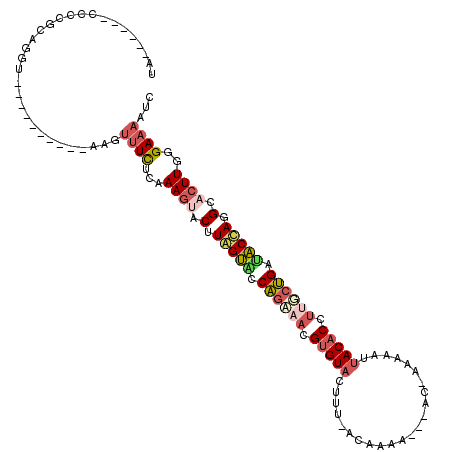
| Location | 16,100,167 – 16,100,269 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -23.95 |
| Consensus MFE | -12.17 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16100167 102 + 20766785 UC------CUACGCAGGU----------AACUUUCUCGAAGUACUUAGUACCAGAAACGUGUACUUU-ACAUAAAC-AUAAAAAAUAACACUUUGCUGAUACCAGGCACUUGGGAAAAUC .(------((....))).----------...((((((.((((.(((.(((.(((....((((.....-........-..........))))....))).))).))).))))))))))... ( -18.00) >DroSec_CAF1 22074 99 + 1 UA------CCCCGCAGGU----------AAGUUUCUCAAAGGGCUUGGUUCCAGAAACGUGUACUAU-ACAA--AC-AC-AAAAAUUACACCUAUCUGAUACCAGGCACUUCCGAAAAUC ((------((.....)))----------)..((((...(((.(((((((..((((...(((((....-....--..-..-......)))))...))))..))))))).)))..))))... ( -24.27) >DroSim_CAF1 22156 96 + 1 UA------CCCCGCAGGU----------AAGUUUCUCAAAGGGCUUGGUUCCAAAAAAGGGUACUAU-ACAA------C-AAAAAUUACACCUCUCUGAAACCAGGCACUUGCGAAAAUC ..------...(((((((----------......((....))((((((((.((....((((((....-....------.-......))).)))...)).)))))))))))))))...... ( -23.06) >DroEre_CAF1 24050 99 + 1 UU------CUCCGUAGUC----------AUGUUUCUCAAAGUACCUAGUACCAGUAACGUGUACUUU-ACCCAA---AU-AAAAAUUACACCUUGCUGAUACUAUGCACUUGGGAAAAUC ..------..........----------...((((((((.((.(.(((((.((((((.(((((.(((-......---..-..))).))))).)))))).))))).).))))))))))... ( -24.80) >DroYak_CAF1 21775 99 + 1 UC------CUCCGUAGUU----------AGGUUUCUCGAAGUACGUAGUACCAGUAACGUGUACUUU-AACCAA---AC-AAAAAUUACACCUUGCUGAUACUAAGCACUUGGGAAAAUC .(------((........----------)))((((((.((((.(.(((((.((((((.(((((.(((-......---..-..))).))))).)))))).))))).).))))))))))... ( -25.30) >DroAna_CAF1 22081 119 + 1 UAAAAUAUUCACACACUUAAUUCAGAAGAAUUGUUUCAAAAUACAUAGAGUCGGAACCGUGUACUUGUGCGAGAAUAAG-ACAAGAUACACGAUGCCGGUUCUAAGAAAUUAUGAACAAC .......((((......((((((....))))))............((((..(((...(((((((((((.(........)-))))).))))))...)))..))))........)))).... ( -28.30) >consensus UA______CCCCGCAGGU__________AAGUUUCUCAAAGUACUUAGUACCAGAAACGUGUACUUU_ACAAAA___AC_AAAAAUUACACCUUGCUGAUACCAGGCACUUGGGAAAAUC ...............................((((...((((.(.(((((.((((((.(((((.......................))))).)))))).))))).).))))..))))... (-12.17 = -13.12 + 0.95)

| Location | 16,100,167 – 16,100,269 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.42 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -20.18 |
| Energy contribution | -20.07 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16100167 102 - 20766785 GAUUUUCCCAAGUGCCUGGUAUCAGCAAAGUGUUAUUUUUUAU-GUUUAUGU-AAAGUACACGUUUCUGGUACUAAGUACUUCGAGAAAGUU----------ACCUGCGUAG------GA ((((((((.((((((.(((((((((....((((....((((((-......))-)))).))))....))))))))).)))))).).)))))))----------.(((....))------). ( -31.40) >DroSec_CAF1 22074 99 - 1 GAUUUUCGGAAGUGCCUGGUAUCAGAUAGGUGUAAUUUUU-GU-GU--UUGU-AUAGUACACGUUUCUGGAACCAAGCCCUUUGAGAAACUU----------ACCUGCGGGG------UA ..((((((((.(.((.((((.(((((.(.(((((....((-((-(.--...)-))))))))).).))))).)))).))).))))))))...(----------(((.....))------)) ( -27.90) >DroSim_CAF1 22156 96 - 1 GAUUUUCGCAAGUGCCUGGUUUCAGAGAGGUGUAAUUUUU-G------UUGU-AUAGUACCCUUUUUUGGAACCAAGCCCUUUGAGAAACUU----------ACCUGCGGGG------UA ..((((((.(((.((.((((((((((((((.(((......-.------....-....))).)))))))))))))).)).)))))))))...(----------(((.....))------)) ( -28.46) >DroEre_CAF1 24050 99 - 1 GAUUUUCCCAAGUGCAUAGUAUCAGCAAGGUGUAAUUUUU-AU---UUGGGU-AAAGUACACGUUACUGGUACUAGGUACUUUGAGAAACAU----------GACUACGGAG------AA ..(((((..((((((.(((((((((.((.(((((...(((-((---....))-))).))))).)).))))))))).)))))).)))))....----------..(....)..------.. ( -29.80) >DroYak_CAF1 21775 99 - 1 GAUUUUCCCAAGUGCUUAGUAUCAGCAAGGUGUAAUUUUU-GU---UUGGUU-AAAGUACACGUUACUGGUACUACGUACUUCGAGAAACCU----------AACUACGGAG------GA ...(((((.((((((.(((((((((.((.(((((...(((-(.---.....)-))).))))).)).))))))))).)))))).).))))(((----------..(....)))------). ( -30.90) >DroAna_CAF1 22081 119 - 1 GUUGUUCAUAAUUUCUUAGAACCGGCAUCGUGUAUCUUGU-CUUAUUCUCGCACAAGUACACGGUUCCGACUCUAUGUAUUUUGAAACAAUUCUUCUGAAUUAAGUGUGUGAAUAUUUUA ..((((((((......((((..(((.((((((((.(((((-(........).))))))))))))).)))..)))).............(((((....))))).....))))))))..... ( -30.80) >consensus GAUUUUCCCAAGUGCCUAGUAUCAGCAAGGUGUAAUUUUU_GU___UUUUGU_AAAGUACACGUUUCUGGUACUAAGUACUUUGAGAAACUU__________AACUGCGGAG______UA ..(((((...(((((.(((((((((.((.(((((.((((..............))))))))).)).))))))))).)))))..)))))................................ (-20.18 = -20.07 + -0.10)
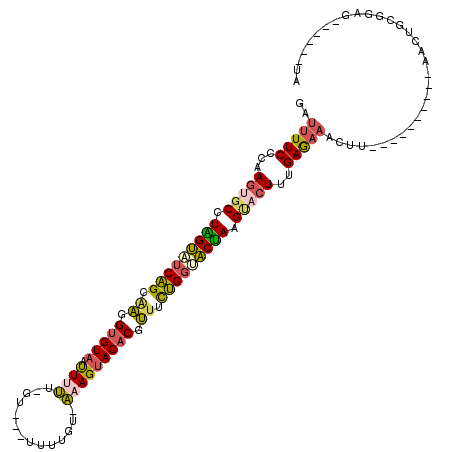
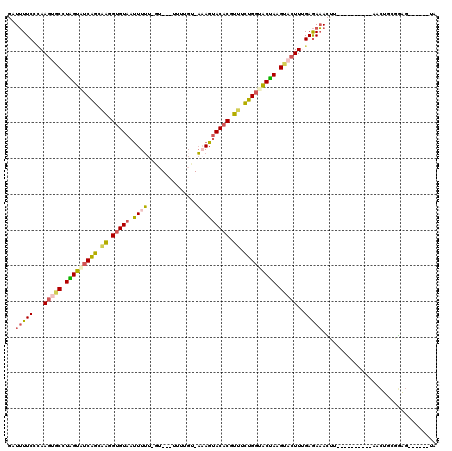
| Location | 16,100,191 – 16,100,303 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.59 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -10.38 |
| Energy contribution | -10.63 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16100191 112 + 20766785 GUACUUAGUACCAGAAACGUGUACUUU-ACAUAAAC-AUAAAAAAUAACACUUUGCUGAUACCAGGCACUUGGGAAAAUCAUGUCU------AGAUUUGUCAACAUUUUAAAUGGCUUAC (((...((((((......).))))).)-))......-.............((..((((((.((((....))))....)))).))..------))....((((..........)))).... ( -13.40) >DroSec_CAF1 22098 109 + 1 GGGCUUGGUUCCAGAAACGUGUACUAU-ACAA--AC-AC-AAAAAUUACACCUAUCUGAUACCAGGCACUUCCGAAAAUCAUGUCU------AGAUUCCUCAACAUUUUAAUUGCCUUAC (((((((((..((((...(((((....-....--..-..-......)))))...))))..))))))).............((((..------((....))..))))........)).... ( -21.87) >DroSim_CAF1 22180 106 + 1 GGGCUUGGUUCCAAAAAAGGGUACUAU-ACAA------C-AAAAAUUACACCUCUCUGAAACCAGGCACUUGCGAAAAUCAUGUCU------AGAUUCCUCAACAUUUUAAUUGCCUUAC ..((((((((.((....((((((....-....------.-......))).)))...)).))))))))....((((.....((((..------((....))..)))).....))))..... ( -19.26) >DroEre_CAF1 24074 102 + 1 GUACCUAGUACCAGUAACGUGUACUUU-ACCCAA---AU-AAAAAUUACACCUUGCUGAUACUAUGCACUUGGGAAAAUCAG-------------GUUCUAGACAUCUUAACUUAUUUAC (((..(((((.((((((.(((((.(((-......---..-..))).))))).)))))).))))))))((((((.....))))-------------))....................... ( -20.30) >DroYak_CAF1 21799 110 + 1 GUACGUAGUACCAGUAACGUGUACUUU-AACCAA---AC-AAAAAUUACACCUUGCUGAUACUAAGCACUUGGGAAAAUCAGAUCC-----AAGAGUCCUGGACAUCGGAAGCCACUUAC .....(((((.((((((.(((((.(((-......---..-..))).))))).)))))).))))).((.((((((.........)))-----)))..(((........))).))....... ( -24.30) >DroAna_CAF1 22121 113 + 1 AUACAUAGAGUCGGAACCGUGUACUUGUGCGAGAAUAAG-ACAAGAUACACGAUGCCGGUUCUAAGAAAUUAUGAACAACUUUUUUUUUCUUUAAUUCCAAGA------AAUCACUUUAC .....(((((((((...(((((((((((.(........)-))))).))))))...)))(((((((....))).)))).........((((((.......))))------))..)))))). ( -26.90) >consensus GUACUUAGUACCAGAAACGUGUACUUU_ACAAAA___AC_AAAAAUUACACCUUGCUGAUACCAGGCACUUGGGAAAAUCAUGUCU______AGAUUCCUCAACAUUUUAAUUGCCUUAC .....(((((.((((((.(((((.......................))))).)))))).)))))........................................................ (-10.38 = -10.63 + 0.25)

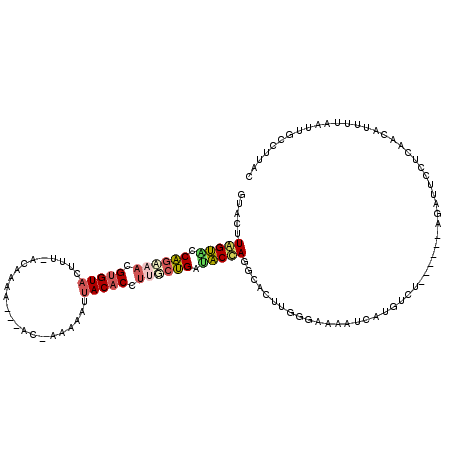
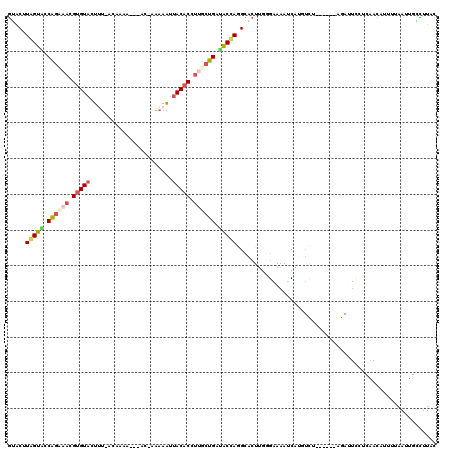
| Location | 16,100,191 – 16,100,303 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.59 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -16.41 |
| Energy contribution | -16.11 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16100191 112 - 20766785 GUAAGCCAUUUAAAAUGUUGACAAAUCU------AGACAUGAUUUUCCCAAGUGCCUGGUAUCAGCAAAGUGUUAUUUUUUAU-GUUUAUGU-AAAGUACACGUUUCUGGUACUAAGUAC .................(((..(((((.------......)))))...)))((((.(((((((((....((((....((((((-......))-)))).))))....))))))))).)))) ( -22.80) >DroSec_CAF1 22098 109 - 1 GUAAGGCAAUUAAAAUGUUGAGGAAUCU------AGACAUGAUUUUCGGAAGUGCCUGGUAUCAGAUAGGUGUAAUUUUU-GU-GU--UUGU-AUAGUACACGUUUCUGGAACCAAGCCC ...(((((......(((((.((....))------.)))))......(....))))))(((.(((((.(.(((((....((-((-(.--...)-))))))))).).))))).)))...... ( -25.90) >DroSim_CAF1 22180 106 - 1 GUAAGGCAAUUAAAAUGUUGAGGAAUCU------AGACAUGAUUUUCGCAAGUGCCUGGUUUCAGAGAGGUGUAAUUUUU-G------UUGU-AUAGUACCCUUUUUUGGAACCAAGCCC ...(((((......(((((.((....))------.)))))......(....))))))(((((((((((((.(((......-.------....-....))).)))))))))))))...... ( -26.66) >DroEre_CAF1 24074 102 - 1 GUAAAUAAGUUAAGAUGUCUAGAAC-------------CUGAUUUUCCCAAGUGCAUAGUAUCAGCAAGGUGUAAUUUUU-AU---UUGGGU-AAAGUACACGUUACUGGUACUAGGUAC .............((.(((......-------------..)))..))....((((.(((((((((.((.(((((...(((-((---....))-))).))))).)).))))))))).)))) ( -24.80) >DroYak_CAF1 21799 110 - 1 GUAAGUGGCUUCCGAUGUCCAGGACUCUU-----GGAUCUGAUUUUCCCAAGUGCUUAGUAUCAGCAAGGUGUAAUUUUU-GU---UUGGUU-AAAGUACACGUUACUGGUACUACGUAC .....(((........(((((((...)))-----)))).........))).((((.(((((((((.((.(((((...(((-(.---.....)-))).))))).)).))))))))).)))) ( -30.73) >DroAna_CAF1 22121 113 - 1 GUAAAGUGAUU------UCUUGGAAUUAAAGAAAAAAAAAGUUGUUCAUAAUUUCUUAGAACCGGCAUCGUGUAUCUUGU-CUUAUUCUCGCACAAGUACACGGUUCCGACUCUAUGUAU .........((------((((.......))))))......................((((..(((.((((((((.(((((-(........).))))))))))))).)))..))))..... ( -25.50) >consensus GUAAGGCAAUUAAAAUGUCGAGGAAUCU______AGACAUGAUUUUCCCAAGUGCCUAGUAUCAGCAAGGUGUAAUUUUU_GU___UUUUGU_AAAGUACACGUUUCUGGUACUAAGUAC ...................................................((((.(((((((((.((.(((((.((((..............))))))))).)).))))))))).)))) (-16.41 = -16.11 + -0.30)
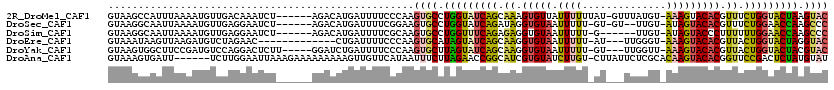

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:43 2006