| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,100,047 – 16,100,154 |
| Length | 107 |
| Max. P | 0.997009 |

| Location | 16,100,047 – 16,100,154 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
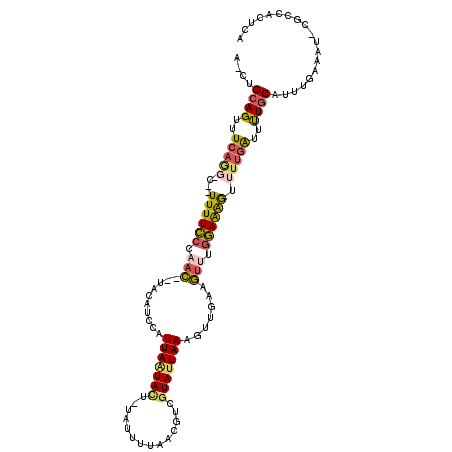
| Mean single sequence MFE | -25.09 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.55 |
| Covariance contribution | 0.03 |
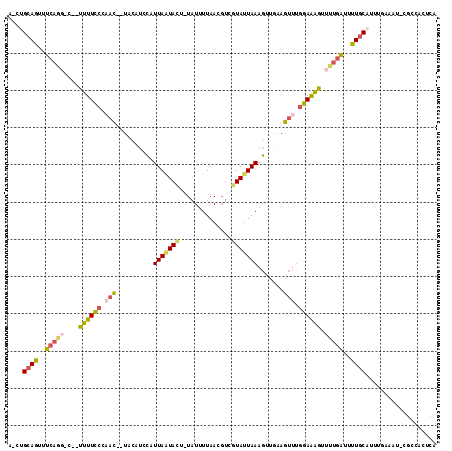
| Combinations/Pair | 1.40 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
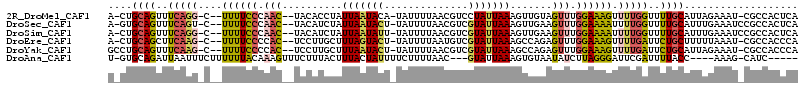
>2R_DroMel_CAF1 16100047 107 - 20766785 A-CUGCAGUUUCAGG-C--UUUUCCCAAC--UACACCUAUUAAUACA-UAUUUUAACGUCCUAUUAAAGUUGUAGUUUGGAAAGUUUUGGUUUUGCAUUAGAAAU-CGCCACUCA .-.(((((..(((((-.--((((((.(((--((((.((.((((((..-.............)))))))).))))))).)))))).)))))..)))))........-......... ( -25.46) >DroSec_CAF1 21953 108 - 1 A-GUGCAGUUUCAGU-C--UUUUCCCAAC--UACAUCUAUUAAUACU-UAUUUUAACGUCGUAUUAAAGUUGAAGUUUGGAAAAUUUUGGUUUUGCAUUUGAAAUCCGCCACUCA (-((((((..((((.-.--((((((.(((--(.((.((.(((((((.-............))))))))).)).)))).))))))..))))..)))))))................ ( -26.02) >DroSim_CAF1 22035 108 - 1 A-CUGCAGUUUCAGG-C--UUUUCCCAAC--UACAUCUAUUAAUAUU-UAUUUUAACGUCGUAUUAAAGUUGAAGUUUGGAAAAUUUUGGUUUUGCAUUUGAAAUCCGCCACUCA .-.(((((..(((((-.--((((((.(((--(.((.((.(((((((.-............))))))))).)).)))).)))))).)))))..))))).................. ( -22.72) >DroEre_CAF1 23931 107 - 1 A-CUGCAGCUUCAAG-C--UUUUCCCCAC--UCCUUGCUUUAGUACU-UAUUUUAAUGUCGUAUUAAAGCCAGAGUUUGGAAAGUUUUGAUUCUGCUUUUUAAAU-CGCCACCCA .-..((((..(((((-.--((((((..((--((...((((((((((.-............))))))))))..))))..)))))).)))))..)))).........-......... ( -31.42) >DroYak_CAF1 21655 108 - 1 GCCUGCAGUUUCAAG-C--UUUUCCCCAC--UCCUUGCUUUAAUACU-UAUUUUAACGUCGUAUUAAAGCCAGAGUUUGGAAAGUUUUGAUUCUGCAUUAGAAAU-CGCCACCCA ...(((((..(((((-.--((((((..((--((...((((((((((.-............))))))))))..))))..)))))).)))))..)))))........-......... ( -33.32) >DroAna_CAF1 21940 101 - 1 U-GUGCAGAUUAAUUUCUUUUUUACAAAGUUUCUUUACUUUACUAUUUUCUUUUAAC---GUAUUAAAGUGUAAUAUCUUAGGGAUUCGAUUUUACC----AAAG-CAUC----- .-((((((((......((((..((((((((......)))))..((........))..---)))..))))......))))..((............))----...)-))).----- ( -11.60) >consensus A_CUGCAGUUUCAGG_C__UUUUCCCAAC__UACAUCCAUUAAUACU_UAUUUUAACGUCGUAUUAAAGUUGAAGUUUGGAAAGUUUUGAUUUUGCAUUUGAAAU_CGCCACUCA ....((((..(((((....((((((.(((..........(((((((..............))))))).......))).)))))).)))))..))))................... (-10.52 = -10.55 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:39 2006