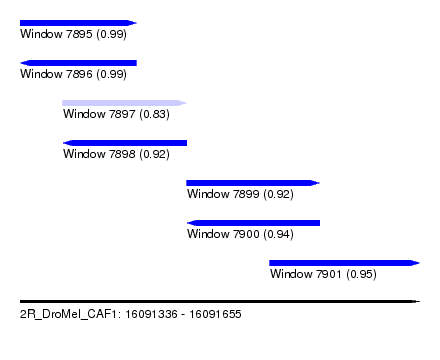
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 16,091,336 – 16,091,655 |
| Length | 319 |
| Max. P | 0.994938 |

| Location | 16,091,336 – 16,091,429 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -23.69 |
| Consensus MFE | -15.10 |
| Energy contribution | -16.43 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091336 93 + 20766785 GGCCUCAUAAAUAUGUUUGGCCAU---GCCUUUGGGCCUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUCU------GUCUCCGUUUUUUU--------UUCAACUUUU ((((.(((....)))...))))..---(((....)))..(((((((((..((.....))...))))))))).------..............--------.......... ( -26.40) >DroSec_CAF1 12785 92 + 1 GGCCUCAUAAAUAUGUUUGGUCAU---GCCUUUGGGCCUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUGU------GUCUCCGUUUUAUU--------UUCAACUU-U ((((.((....((((......)))---)....)))))).(((((((..(((..(((.....)))..))).))------))))).........--------........-. ( -22.60) >DroSim_CAF1 13058 92 + 1 GGCCUCAUAAAUAUGUUUGGCCAU---GCCUUUGGGCCUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUCU------GUCUCCGUUUUAUU--------UUCAACUU-U ((((.(((....)))...))))..---(((....)))..(((((((((..((.....))...))))))))).------..............--------........-. ( -26.40) >DroEre_CAF1 13023 91 + 1 GGCCUCAUAAAUAUGUUUGGCCAU---GCCUUUGGGCCUGAGACGCUUGACAAUUUCUGCAA-AGCGUCUCU------GUCUCCGUUGUAUU--------UUUAUUUU-U ((((.(((....)))...))))((---((...((((...(((((((((..((.....))..)-)))))))).------...))))..)))).--------........-. ( -28.30) >DroWil_CAF1 26992 76 + 1 AGCCUCAUAAAUAUAUUGGCCUAAGGAGUUUUUUUGCCGACGACAGUUGACAAUUUUUUU--------------------CUUCUCU-------------UUUUUUUU-U .(((.............)))...(((((.....(((.((((....)))).))).......--------------------)))))..-------------........-. ( -10.92) >DroYak_CAF1 13342 105 + 1 GGCCUCAUAAAUAUGUUUGGCCAU---GCCGUUGGGCCUGAGACGCUUGACAAUUUCUGCAA-AGCGUCUCUGUCUCUGUCUCCGUUGUAUUUCAAUUUUUUUUUUUU-U ((((.(((....)))...))))..---(((....)))..(((((((((..((.....))..)-)))))))).............((((.....))))...........-. ( -27.50) >consensus GGCCUCAUAAAUAUGUUUGGCCAU___GCCUUUGGGCCUGAGACGCUUGACAAUUUCUGCAA_AGCGUCUCU______GUCUCCGUUUUAUU________UUCAACUU_U ((((..(((....)))..)))).....(((....)))..((((((((((.((.....))))..))))))))....................................... (-15.10 = -16.43 + 1.34)

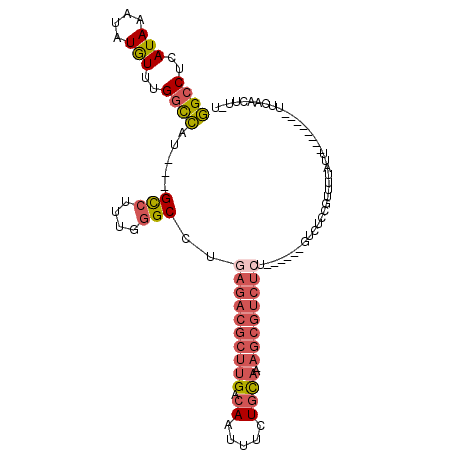
| Location | 16,091,336 – 16,091,429 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 75.94 |
| Mean single sequence MFE | -23.75 |
| Consensus MFE | -14.29 |
| Energy contribution | -16.07 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.53 |
| SVM RNA-class probability | 0.994938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091336 93 - 20766785 AAAAGUUGAA--------AAAAAAACGGAGAC------AGAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAGGCCCAAAGGC---AUGGCCAAACAUAUUUAUGAGGCC ....(((...--------.....)))(....)------.(((((((((..((((...)))).))))))))).((((((.(((.---(((......))).))).)).)))) ( -27.40) >DroSec_CAF1 12785 92 - 1 A-AAGUUGAA--------AAUAAAACGGAGAC------ACAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAGGCCCAAAGGC---AUGACCAAACAUAUUUAUGAGGCC .-..(((...--------.....)))(....)------..((((((((..((((...)))).))))))))..((((((.(((.---(((......))).))).)).)))) ( -21.70) >DroSim_CAF1 13058 92 - 1 A-AAGUUGAA--------AAUAAAACGGAGAC------AGAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAGGCCCAAAGGC---AUGGCCAAACAUAUUUAUGAGGCC .-..(((...--------.....)))(....)------.(((((((((..((((...)))).))))))))).((((((.(((.---(((......))).))).)).)))) ( -27.40) >DroEre_CAF1 13023 91 - 1 A-AAAAUAAA--------AAUACAACGGAGAC------AGAGACGCU-UUGCAGAAAUUGUCAAGCGUCUCAGGCCCAAAGGC---AUGGCCAAACAUAUUUAUGAGGCC .-........--------........(....)------.((((((((-(.((((...)))).))))))))).((((((.(((.---(((......))).))).)).)))) ( -28.60) >DroWil_CAF1 26992 76 - 1 A-AAAAAAAA-------------AGAGAAG--------------------AAAAAAAUUGUCAACUGUCGUCGGCAAAAAAACUCCUUAGGCCAAUAUAUUUAUGAGGCU .-........-------------.......--------------------.......(((((.((....)).)))))............((((.(((....)))..)))) ( -9.20) >DroYak_CAF1 13342 105 - 1 A-AAAAAAAAAAAAUUGAAAUACAACGGAGACAGAGACAGAGACGCU-UUGCAGAAAUUGUCAAGCGUCUCAGGCCCAACGGC---AUGGCCAAACAUAUUUAUGAGGCC .-........................(....).......((((((((-(.((((...)))).))))))))).((((....(((---...)))...(((....))).)))) ( -28.20) >consensus A_AAAAUAAA________AAUAAAACGGAGAC______AGAGACGCU_UUGCAGAAAUUGUCAAGCGUCUCAGGCCCAAAGGC___AUGGCCAAACAUAUUUAUGAGGCC .......................................((((((((...((((...))))..)))))))).((((....(((......)))...(((....))).)))) (-14.29 = -16.07 + 1.78)


| Location | 16,091,370 – 16,091,469 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -21.22 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091370 99 + 20766785 CUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUCU------GUCUCCGUUUUUUU--------UUCAACUUUUUUGCGCACUUUUUGGUGAGAAAUGGGAGCGGAAAUUAAUG ..(((((((((..((.....))...))))))))).------...((((((((..(--------(((..(......)..((((....)))).))))..))))))))........ ( -28.80) >DroSec_CAF1 12819 97 + 1 CUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUGU------GUCUCCGUUUUAUU--------UUCAACUU-UUUGCGCACUU-UUGGUGAGAAAUGGGAGCGGAAAUUAAUG ...((((((((..((.....))...))))))))..------...((((((((.((--------((((.((.-...........-..))))))))...))))))))........ ( -23.34) >DroSim_CAF1 13092 97 + 1 CUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUCU------GUCUCCGUUUUAUU--------UUCAACUU-UUUGCGCACUU-UUGGUGAGAAAUGGGAGCGGAAAUUAAUG ..(((((((((..((.....))...))))))))).------...((((((((.((--------((((.((.-...........-..))))))))...))))))))........ ( -26.34) >DroEre_CAF1 13057 96 + 1 CUGAGACGCUUGACAAUUUCUGCAA-AGCGUCUCU------GUCUCCGUUGUAUU--------UUUAUUUU-UUUGCGCACUU-UUGGUGAGAAAUGGGAGCGGAAAUUAAUG ..(((((((((..((.....))..)-)))))))).------...((((((.(...--------.....(((-((..(.((...-.)))..)))))..).))))))........ ( -26.00) >DroYak_CAF1 13376 111 + 1 CUGAGACGCUUGACAAUUUCUGCAA-AGCGUCUCUGUCUCUGUCUCCGUUGUAUUUCAAUUUUUUUUUUUU-UUUGCGCACUUUUUGGUGAGAAAUGGGAGCGGAAAUUAAUG ..(((((((((..((.....))..)-))))))))....((((.((((((((.....))))........(((-((..(.((.....)))..)))))..))))))))........ ( -30.40) >consensus CUGAGACGCUUGACAAUUUCUGCAAAAGCGUCUCU______GUCUCCGUUUUAUU________UUCAACUU_UUUGCGCACUU_UUGGUGAGAAAUGGGAGCGGAAAUUAAUG ..(((((((((..((.....))...))))))))).......((..(((((((................(......)..((((....)))).)))))))..))........... (-21.22 = -21.98 + 0.76)

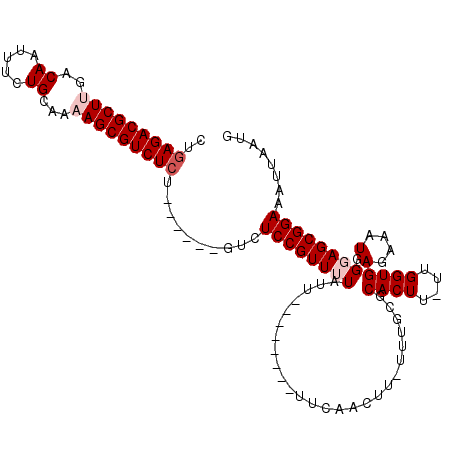
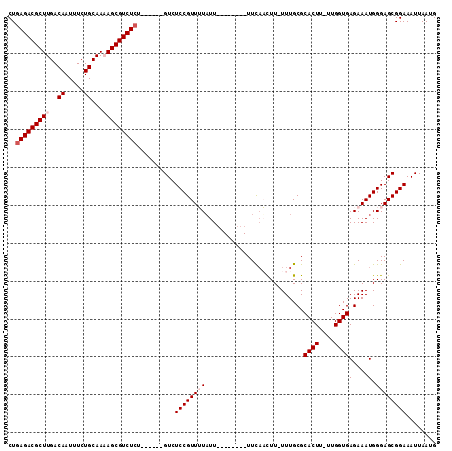
| Location | 16,091,370 – 16,091,469 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 89.58 |
| Mean single sequence MFE | -22.28 |
| Consensus MFE | -17.09 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923740 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091370 99 - 20766785 CAUUAAUUUCCGCUCCCAUUUCUCACCAAAAAGUGCGCAAAAAAGUUGAA--------AAAAAAACGGAGAC------AGAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAG ......(((((((...(((((.........))))).))......(((...--------.....)))))))).------.(((((((((..((((...)))).))))))))).. ( -22.70) >DroSec_CAF1 12819 97 - 1 CAUUAAUUUCCGCUCCCAUUUCUCACCAA-AAGUGCGCAAA-AAGUUGAA--------AAUAAAACGGAGAC------ACAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAG ......(((((((...(((((........-))))).))...-..(((...--------.....)))))))).------..((((((((..((((...)))).))))))))... ( -19.10) >DroSim_CAF1 13092 97 - 1 CAUUAAUUUCCGCUCCCAUUUCUCACCAA-AAGUGCGCAAA-AAGUUGAA--------AAUAAAACGGAGAC------AGAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAG ......(((((((...(((((........-))))).))...-..(((...--------.....)))))))).------.(((((((((..((((...)))).))))))))).. ( -23.50) >DroEre_CAF1 13057 96 - 1 CAUUAAUUUCCGCUCCCAUUUCUCACCAA-AAGUGCGCAAA-AAAAUAAA--------AAUACAACGGAGAC------AGAGACGCU-UUGCAGAAAUUGUCAAGCGUCUCAG ......(((((((...(((((........-))))).))...-...((...--------.)).....))))).------.((((((((-(.((((...)))).))))))))).. ( -21.20) >DroYak_CAF1 13376 111 - 1 CAUUAAUUUCCGCUCCCAUUUCUCACCAAAAAGUGCGCAAA-AAAAAAAAAAAAUUGAAAUACAACGGAGACAGAGACAGAGACGCU-UUGCAGAAAUUGUCAAGCGUCUCAG ......((((.(((((..((((.(((......))).).)))-............(((.....))).)))).).))))..((((((((-(.((((...)))).))))))))).. ( -24.90) >consensus CAUUAAUUUCCGCUCCCAUUUCUCACCAA_AAGUGCGCAAA_AAGUUGAA________AAUAAAACGGAGAC______AGAGACGCUUUUGCAGAAAUUGUCAAGCGUCUCAG ......((((((......((((.(((......))).).)))........................))))))........((((((((...((((...))))..)))))))).. (-17.09 = -17.29 + 0.20)

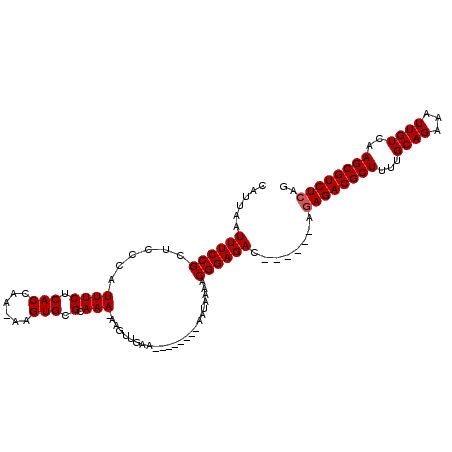
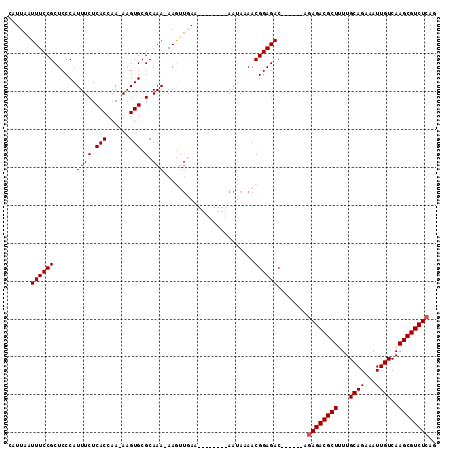
| Location | 16,091,469 – 16,091,575 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -35.98 |
| Consensus MFE | -29.90 |
| Energy contribution | -31.66 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920113 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091469 106 + 20766785 ACCGGAGUGCUGGAAUCUUUGGCAUGCCACUGUUGGCUGUGGCAUCAGGUGCUUUUGCUCUGGAGCGUUUUGCGAAAGUGGUUGGCAGAUUGAAGAGCGGCCAAAA .((((....))))....(((((((((((((........)))))))..........((((((....((.(((((.((.....)).))))).)).)))))))))))). ( -36.50) >DroSec_CAF1 12916 105 + 1 ACCGGAGUGCUGGAAUCUUUGGCAUGCCACAG-UGGCUGUGGCAUCAGGUGCUUUUGCUGCGGAGCGUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGCGGCCAAAA .((((....))))....(((((((((((((((-...)))))))))....(((((((.(((((((.(.....((....))).)))))))....))))))))))))). ( -43.50) >DroSim_CAF1 13189 105 + 1 ACCGGAGUGCUGGAAUCUUUGGCAUGCCACAG-UGGCUGUGGCAUCAGGUGCUUUUGCUGCGGAGCGUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAA .((((....))))....(((((((((((((((-...))))))))).....((((((.(((((((.(.....((....))).)))))))....)))))).)))))). ( -41.10) >DroEre_CAF1 13153 97 + 1 ACCGAAGUGCUGGAAUCUUUGGCAUGCCAG---------UGGCAUCAGGUGGUUUUGCUCUACAGCGUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAA (((...((((..((...))..))))(((..---------.)))....)))((((..(((((.(((((..(..(....)..)..)))....)).))))))))).... ( -31.20) >DroYak_CAF1 13487 97 + 1 ACCGAAGUGCUGGAAUCUUUGGCAUGCCAC---------UGGCAUCAGGUGGUUUUGCUAUGGAGCGUUUUGCGAAAGUGGUUCCCAGAUUGAAGAGUGGCCAAAA .(((.(((.((((..(((.(((((.(((((---------(.......))))))..))))).)))..(..(..(....)..)..))))))))......)))...... ( -27.60) >consensus ACCGGAGUGCUGGAAUCUUUGGCAUGCCAC_G_UGGCUGUGGCAUCAGGUGCUUUUGCUCUGGAGCGUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAA .((((....))))....(((((((((((((((....))))))))).....(((((((((....)))..((((((((.....))))))))...)))))).)))))). (-29.90 = -31.66 + 1.76)



| Location | 16,091,469 – 16,091,575 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.87 |
| Mean single sequence MFE | -25.42 |
| Consensus MFE | -23.00 |
| Energy contribution | -24.20 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091469 106 - 20766785 UUUUGGCCGCUCUUCAAUCUGCCAACCACUUUCGCAAAACGCUCCAGAGCAAAAGCACCUGAUGCCACAGCCAACAGUGGCAUGCCAAAGAUUCCAGCACUCCGGU (((((((.(((........(((.((.....)).)))....(((....)))...))).....(((((((........))))))))))))))................ ( -24.60) >DroSec_CAF1 12916 105 - 1 UUUUGGCCGCUCUUCAAUCUGCGAACCACUUUCGCAAAACGCUCCGCAGCAAAAGCACCUGAUGCCACAGCCA-CUGUGGCAUGCCAAAGAUUCCAGCACUCCGGU (((((((.(((........((((((.....))))))....(((....)))...))).....(((((((((...-))))))))))))))))................ ( -32.00) >DroSim_CAF1 13189 105 - 1 UUUUGGCCACUCUUCAAUCUGCGAACCACUUUCGCAAAACGCUCCGCAGCAAAAGCACCUGAUGCCACAGCCA-CUGUGGCAUGCCAAAGAUUCCAGCACUCCGGU (((((((............((((((.....))))))....(((....)))...........(((((((((...-))))))))))))))))................ ( -28.80) >DroEre_CAF1 13153 97 - 1 UUUUGGCCACUCUUCAAUCUGCGAACCACUUUCGCAAAACGCUGUAGAGCAAAACCACCUGAUGCCA---------CUGGCAUGCCAAAGAUUCCAGCACUUCGGU (((((((..((((.((...((((((.....))))))......)).))))............(((((.---------..))))))))))))........((....)) ( -22.70) >DroYak_CAF1 13487 97 - 1 UUUUGGCCACUCUUCAAUCUGGGAACCACUUUCGCAAAACGCUCCAUAGCAAAACCACCUGAUGCCA---------GUGGCAUGCCAAAGAUUCCAGCACUUCGGU ..................((((((....((((.(((....(((....)))....((((.((....))---------))))..))).)))).)))))).((....)) ( -19.00) >consensus UUUUGGCCACUCUUCAAUCUGCGAACCACUUUCGCAAAACGCUCCACAGCAAAAGCACCUGAUGCCACAGCCA_C_GUGGCAUGCCAAAGAUUCCAGCACUCCGGU (((((((............((((((.....))))))....(((....)))...........(((((((((....))))))))))))))))................ (-23.00 = -24.20 + 1.20)

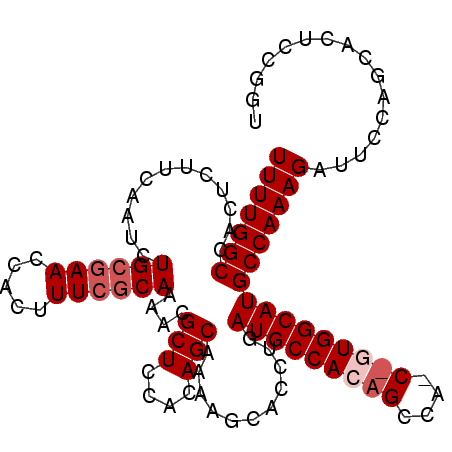
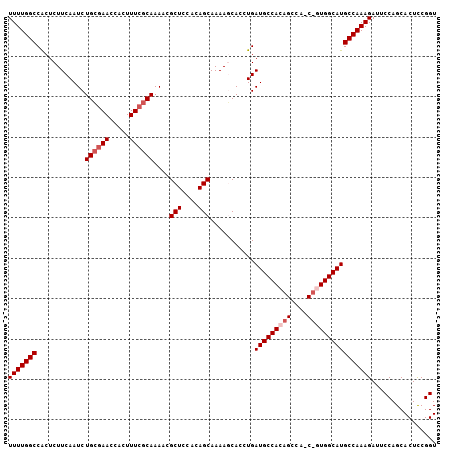
| Location | 16,091,535 – 16,091,655 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -28.26 |
| Energy contribution | -28.98 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 16091535 120 + 20766785 GUUUUGCGAAAGUGGUUGGCAGAUUGAAGAGCGGCCAAAAGAUAAGUAGUUGAUCGGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUGGCCGAGACCUGUUUCGCAAAAGUUAUAAC .((((((((((..(((((((.(((((((((..(((((...((((.(.....((((...)))).).)))).....))))).)))))).))).)))..))))..))))))))))........ ( -35.80) >DroSec_CAF1 12981 120 + 1 GUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGCGGCCAAAAGAUAAGUAGUUGAUCGGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUGGCCAAGACCUGUUUUGCAAAAGUUAUAAC .((((((((((..((((.((..........))((((((.....((((((((((((...))))......(....).)))))))).....))))))..))))..))))))))))........ ( -32.80) >DroSim_CAF1 13254 120 + 1 GUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAAGAUAAGUAGUUGAUCGGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUGGCCGAGACCUGUUUUGCAAAAGUUAUAAC .((((((((((..(((((((......(((((((((((...((((.(.....((((...)))).).)))).....)))))))))))......).))).)))..))))))))))........ ( -33.70) >DroEre_CAF1 13210 120 + 1 GUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAAGAUAAGUAGUUGAUCAGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUUUUCAAGACCUGUUUUACAGAAGUUAUACC ..((((((((.....))))))))...(((((((((((...((((.(.....((((...)))).).)))).....)))))))))))..............(((.....))).......... ( -26.80) >DroYak_CAF1 13544 120 + 1 GUUUUGCGAAAGUGGUUCCCAGAUUGAAGAGUGGCCAAAAGAUAAGUAGUUGAUCAGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUUACCAAGACCUGUUUUAUAGAACUUAUACC ((((((..(((..((((...((((..(((((((((((...((((.(.....((((...)))).).)))).....)))))))))))..)))).....))))..)))..))))))....... ( -26.40) >consensus GUUUUGCGAAAGUGGUUCGCAGAUUGAAGAGUGGCCAAAAGAUAAGUAGUUGAUCGGAGAUCUCAUGUCGAAAGUGGCUAUUUUUAUAUUGGCCAAGACCUGUUUUGCAAAAGUUAUAAC .((((((((((..((((.((......(((((((((((...((((.(.....((((...)))).).)))).....)))))))))))......))...))))..))))))))))........ (-28.26 = -28.98 + 0.72)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:32:32 2006